Torkan Gholamalizadeh
Tooth-Diffusion: Guided 3D CBCT Synthesis with Fine-Grained Tooth Conditioning
Aug 19, 2025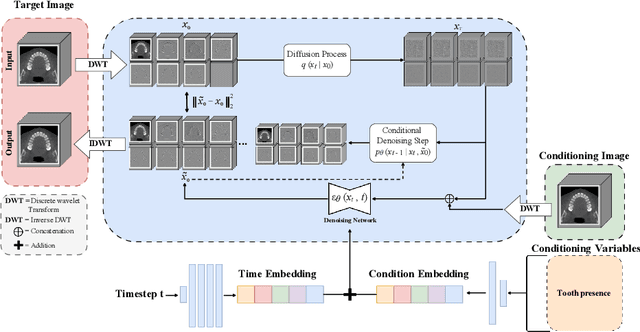
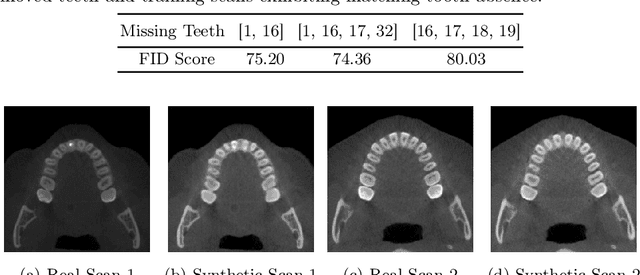
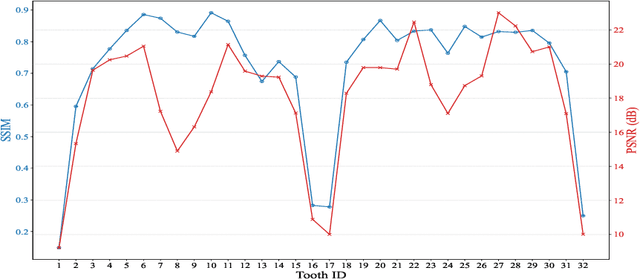
Abstract:Despite the growing importance of dental CBCT scans for diagnosis and treatment planning, generating anatomically realistic scans with fine-grained control remains a challenge in medical image synthesis. In this work, we propose a novel conditional diffusion framework for 3D dental volume generation, guided by tooth-level binary attributes that allow precise control over tooth presence and configuration. Our approach integrates wavelet-based denoising diffusion, FiLM conditioning, and masked loss functions to focus learning on relevant anatomical structures. We evaluate the model across diverse tasks, such as tooth addition, removal, and full dentition synthesis, using both paired and distributional similarity metrics. Results show strong fidelity and generalization with low FID scores, robust inpainting performance, and SSIM values above 0.91 even on unseen scans. By enabling realistic, localized modification of dentition without rescanning, this work opens opportunities for surgical planning, patient communication, and targeted data augmentation in dental AI workflows. The codes are available at: https://github.com/djafar1/tooth-diffusion.
Non-Reference Quality Assessment for Medical Imaging: Application to Synthetic Brain MRIs
Jul 20, 2024



Abstract:Generating high-quality synthetic data is crucial for addressing challenges in medical imaging, such as domain adaptation, data scarcity, and privacy concerns. Existing image quality metrics often rely on reference images, are tailored for group comparisons, or are intended for 2D natural images, limiting their efficacy in complex domains like medical imaging. This study introduces a novel deep learning-based non-reference approach to assess brain MRI quality by training a 3D ResNet. The network is designed to estimate quality across six distinct artifacts commonly encountered in MRI scans. Additionally, a diffusion model is trained on diverse datasets to generate synthetic 3D images of high fidelity. The approach leverages several datasets for training and comprehensive quality assessment, benchmarking against state-of-the-art metrics for real and synthetic images. Results demonstrate superior performance in accurately estimating distortions and reflecting image quality from multiple perspectives. Notably, the method operates without reference images, indicating its applicability for evaluating deep generative models. Besides, the quality scores in the [0, 1] range provide an intuitive assessment of image quality across heterogeneous datasets. Evaluation of generated images offers detailed insights into specific artifacts, guiding strategies for improving generative models to produce high-quality synthetic images. This study presents the first comprehensive method for assessing the quality of real and synthetic 3D medical images in MRI contexts without reliance on reference images.
Auto-segmentation of Hip Joints using MultiPlanar UNet with Transfer learning
Aug 18, 2022
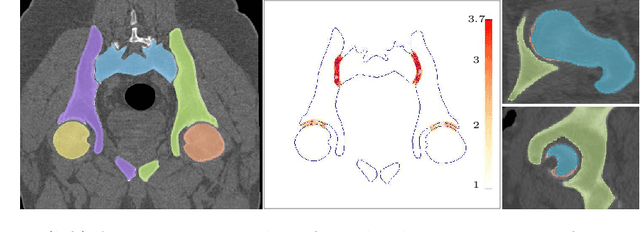


Abstract:Accurate geometry representation is essential in developing finite element models. Although generally good, deep-learning segmentation approaches with only few data have difficulties in accurately segmenting fine features, e.g., gaps and thin structures. Subsequently, segmented geometries need labor-intensive manual modifications to reach a quality where they can be used for simulation purposes. We propose a strategy that uses transfer learning to reuse datasets with poor segmentation combined with an interactive learning step where fine-tuning of the data results in anatomically accurate segmentations suitable for simulations. We use a modified MultiPlanar UNet that is pre-trained using inferior hip joint segmentation combined with a dedicated loss function to learn the gap regions and post-processing to correct tiny inaccuracies on symmetric classes due to rotational invariance. We demonstrate this robust yet conceptually simple approach applied with clinically validated results on publicly available computed tomography scans of hip joints. Code and resulting 3D models are available at: https://github.com/MICCAI2022-155/AuToSeg}
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge