Diego H. Milone
Fairness of Deep Ensembles: On the interplay between per-group task difficulty and under-representation
Jan 24, 2025



Abstract:Ensembling is commonly regarded as an effective way to improve the general performance of models in machine learning, while also increasing the robustness of predictions. When it comes to algorithmic fairness, heterogeneous ensembles, composed of multiple model types, have been employed to mitigate biases in terms of demographic attributes such as sex, age or ethnicity. Moreover, recent work has shown how in multi-class problems even simple homogeneous ensembles may favor performance of the worst-performing target classes. While homogeneous ensembles are simpler to implement in practice, it is not yet clear whether their benefits translate to groups defined not in terms of their target class, but in terms of demographic or protected attributes, hence improving fairness. In this work we show how this simple and straightforward method is indeed able to mitigate disparities, particularly benefiting under-performing subgroups. Interestingly, this can be achieved without sacrificing overall performance, which is a common trade-off observed in bias mitigation strategies. Moreover, we analyzed the interplay between two factors which may result in biases: sub-group under-representation and the inherent difficulty of the task for each group. These results revealed that, contrary to popular assumptions, having balanced datasets may be suboptimal if the task difficulty varies between subgroups. Indeed, we found that a perfectly balanced dataset may hurt both the overall performance and the gap between groups. This highlights the importance of considering the interaction between multiple forces at play in fairness.
Uncertainty in latent representations of variational autoencoders optimized for visual tasks
Apr 23, 2024



Abstract:Deep learning methods are increasingly becoming instrumental as modeling tools in computational neuroscience, employing optimality principles to build bridges between neural responses and perception or behavior. Developing models that adequately represent uncertainty is however challenging for deep learning methods, which often suffer from calibration problems. This constitutes a difficulty in particular when modeling cortical circuits in terms of Bayesian inference, beyond single point estimates such as the posterior mean or the maximum a posteriori. In this work we systematically studied uncertainty representations in latent representations of variational auto-encoders (VAEs), both in a perceptual task from natural images and in two other canonical tasks of computer vision, finding a poor alignment between uncertainty and informativeness or ambiguities in the images. We next showed how a novel approach which we call explaining-away variational auto-encoders (EA-VAEs), fixes these issues, producing meaningful reports of uncertainty in a variety of scenarios, including interpolation, image corruption, and even out-of-distribution detection. We show EA-VAEs may prove useful both as models of perception in computational neuroscience and as inference tools in computer vision.
Multi-view Hybrid Graph Convolutional Network for Volume-to-mesh Reconstruction in Cardiovascular MRI
Nov 22, 2023Abstract:Cardiovascular magnetic resonance imaging is emerging as a crucial tool to examine cardiac morphology and function. Essential to this endeavour are anatomical 3D surface and volumetric meshes derived from CMR images, which facilitate computational anatomy studies, biomarker discovery, and in-silico simulations. However, conventional surface mesh generation methods, such as active shape models and multi-atlas segmentation, are highly time-consuming and require complex processing pipelines to generate simulation-ready 3D meshes. In response, we introduce HybridVNet, a novel architecture for direct image-to-mesh extraction seamlessly integrating standard convolutional neural networks with graph convolutions, which we prove can efficiently handle surface and volumetric meshes by encoding them as graph structures. To further enhance accuracy, we propose a multiview HybridVNet architecture which processes both long axis and short axis CMR, showing that it can increase the performance of cardiac MR mesh generation. Our model combines traditional convolutional networks with variational graph generative models, deep supervision and mesh-specific regularisation. Experiments on a comprehensive dataset from the UK Biobank confirm the potential of HybridVNet to significantly advance cardiac imaging and computational cardiology by efficiently generating high-fidelity and simulation ready meshes from CMR images.
Unsupervised bias discovery in medical image segmentation
Sep 01, 2023Abstract:It has recently been shown that deep learning models for anatomical segmentation in medical images can exhibit biases against certain sub-populations defined in terms of protected attributes like sex or ethnicity. In this context, auditing fairness of deep segmentation models becomes crucial. However, such audit process generally requires access to ground-truth segmentation masks for the target population, which may not always be available, especially when going from development to deployment. Here we propose a new method to anticipate model biases in biomedical image segmentation in the absence of ground-truth annotations. Our unsupervised bias discovery method leverages the reverse classification accuracy framework to estimate segmentation quality. Through numerical experiments in synthetic and realistic scenarios we show how our method is able to successfully anticipate fairness issues in the absence of ground-truth labels, constituting a novel and valuable tool in this field.
CheXmask: a large-scale dataset of anatomical segmentation masks for multi-center chest x-ray images
Jul 06, 2023Abstract:The development of successful artificial intelligence models for chest X-ray analysis relies on large, diverse datasets with high-quality annotations. While several databases of chest X-ray images have been released, most include disease diagnosis labels but lack detailed pixel-level anatomical segmentation labels. To address this gap, we introduce an extensive chest X-ray multi-center segmentation dataset with uniform and fine-grain anatomical annotations for images coming from six well-known publicly available databases: CANDID-PTX, ChestX-ray8, Chexpert, MIMIC-CXR-JPG, Padchest, and VinDr-CXR, resulting in 676,803 segmentation masks. Our methodology utilizes the HybridGNet model to ensure consistent and high-quality segmentations across all datasets. Rigorous validation, including expert physician evaluation and automatic quality control, was conducted to validate the resulting masks. Additionally, we provide individualized quality indices per mask and an overall quality estimation per dataset. This dataset serves as a valuable resource for the broader scientific community, streamlining the development and assessment of innovative methodologies in chest X-ray analysis. The CheXmask dataset is publicly available at: \url{https://physionet.org/content/chexmask-cxr-segmentation-data/}.
Multi-center anatomical segmentation with heterogeneous labels via landmark-based models
Nov 14, 2022Abstract:Learning anatomical segmentation from heterogeneous labels in multi-center datasets is a common situation encountered in clinical scenarios, where certain anatomical structures are only annotated in images coming from particular medical centers, but not in the full database. Here we first show how state-of-the-art pixel-level segmentation models fail in naively learning this task due to domain memorization issues and conflicting labels. We then propose to adopt HybridGNet, a landmark-based segmentation model which learns the available anatomical structures using graph-based representations. By analyzing the latent space learned by both models, we show that HybridGNet naturally learns more domain-invariant feature representations, and provide empirical evidence in the context of chest X-ray multiclass segmentation. We hope these insights will shed light on the training of deep learning models with heterogeneous labels from public and multi-center datasets.
Improving anatomical plausibility in medical image segmentation via hybrid graph neural networks: applications to chest x-ray analysis
Apr 01, 2022

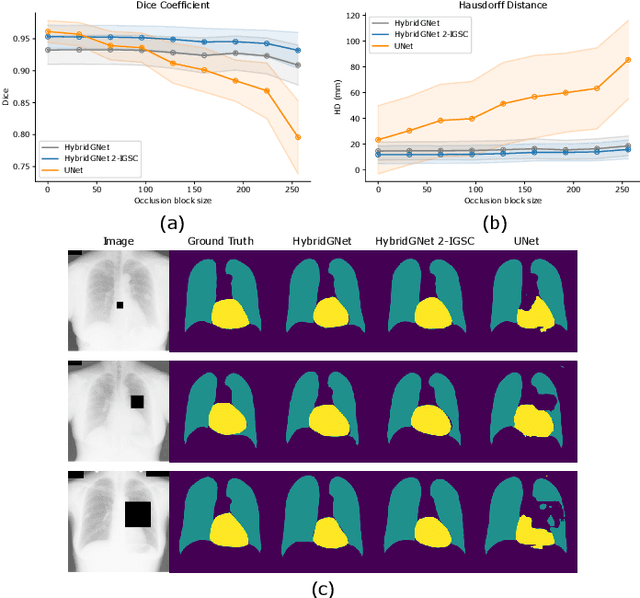

Abstract:Anatomical segmentation is a fundamental task in medical image computing, generally tackled with fully convolutional neural networks which produce dense segmentation masks. These models are often trained with loss functions such as cross-entropy or Dice, which assume pixels to be independent of each other, thus ignoring topological errors and anatomical inconsistencies. We address this limitation by moving from pixel-level to graph representations, which allow to naturally incorporate anatomical constraints by construction. To this end, we introduce HybridGNet, an encoder-decoder neural architecture that leverages standard convolutions for image feature encoding and graph convolutional neural networks (GCNNs) to decode plausible representations of anatomical structures. We also propose a novel image-to-graph skip connection layer which allows localized features to flow from standard convolutional blocks to GCNN blocks, and show that it improves segmentation accuracy. The proposed architecture is extensively evaluated in a variety of domain shift and image occlusion scenarios, and audited considering different types of demographic domain shift. Our comprehensive experimental setup compares HybridGNet with other landmark and pixel-based models for anatomical segmentation in chest x-ray images, and shows that it produces anatomically plausible results in challenging scenarios where other models tend to fail.
Using segment-based features of jaw movements to recognize foraging activities in grazing cattle
Apr 01, 2022



Abstract:Precision livestock farming optimizes livestock production through the use of sensor information and communication technologies to support decision making, proactively and near real-time. Among available technologies to monitor foraging behavior, the acoustic method has been highly reliable and repeatable, but can be subject to further computational improvements to increase precision and specificity of recognition of foraging activities. In this study, an algorithm called Jaw Movement segment-based Foraging Activity Recognizer (JMFAR) is proposed. The method is based on the computation and analysis of temporal, statistical and spectral features of jaw movement sounds for detection of rumination and grazing bouts. They are called JM-segment features because they are extracted from a sound segment and expect to capture JM information of the whole segment rather than individual JMs. Two variants of the method are proposed and tested: (i) the temporal and statistical features only JMFAR-ns; and (ii) a feature selection process (JMFAR-sel). The JMFAR was tested on signals registered in a free grazing environment, achieving an average weighted F1-score greater than 95%. Then, it was compared with a state-of-the-art algorithm, showing improved performance for estimation of grazing bouts (+19%). The JMFAR-ns variant reduced the computational cost by 25.4%, but achieved a slightly lower performance than the JMFAR. The good performance and low computational cost of JMFAR-ns supports the feasibility of using this algorithm variant for real-time implementation in low-cost embedded systems.
Domain Generalization via Gradient Surgery
Aug 03, 2021



Abstract:In real-life applications, machine learning models often face scenarios where there is a change in data distribution between training and test domains. When the aim is to make predictions on distributions different from those seen at training, we incur in a domain generalization problem. Methods to address this issue learn a model using data from multiple source domains, and then apply this model to the unseen target domain. Our hypothesis is that when training with multiple domains, conflicting gradients within each mini-batch contain information specific to the individual domains which is irrelevant to the others, including the test domain. If left untouched, such disagreement may degrade generalization performance. In this work, we characterize the conflicting gradients emerging in domain shift scenarios and devise novel gradient agreement strategies based on gradient surgery to alleviate their effect. We validate our approach in image classification tasks with three multi-domain datasets, showing the value of the proposed agreement strategy in enhancing the generalization capability of deep learning models in domain shift scenarios.
Learning Deformable Registration of Medical Images with Anatomical Constraints
Jan 22, 2020



Abstract:Deformable image registration is a fundamental problem in the field of medical image analysis. During the last years, we have witnessed the advent of deep learning-based image registration methods which achieve state-of-the-art performance, and drastically reduce the required computational time. However, little work has been done regarding how can we encourage our models to produce not only accurate, but also anatomically plausible results, which is still an open question in the field. In this work, we argue that incorporating anatomical priors in the form of global constraints into the learning process of these models, will further improve their performance and boost the realism of the warped images after registration. We learn global non-linear representations of image anatomy using segmentation masks, and employ them to constraint the registration process. The proposed AC-RegNet architecture is evaluated in the context of chest X-ray image registration using three different datasets, where the high anatomical variability makes the task extremely challenging. Our experiments show that the proposed anatomically constrained registration model produces more realistic and accurate results than state-of-the-art methods, demonstrating the potential of this approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge