Candelaria Mosquera
Health Informatics Department Hospital Italiano de Buenos Aires, Universidad Tecnologica Nacional
CheXmask: a large-scale dataset of anatomical segmentation masks for multi-center chest x-ray images
Jul 06, 2023Abstract:The development of successful artificial intelligence models for chest X-ray analysis relies on large, diverse datasets with high-quality annotations. While several databases of chest X-ray images have been released, most include disease diagnosis labels but lack detailed pixel-level anatomical segmentation labels. To address this gap, we introduce an extensive chest X-ray multi-center segmentation dataset with uniform and fine-grain anatomical annotations for images coming from six well-known publicly available databases: CANDID-PTX, ChestX-ray8, Chexpert, MIMIC-CXR-JPG, Padchest, and VinDr-CXR, resulting in 676,803 segmentation masks. Our methodology utilizes the HybridGNet model to ensure consistent and high-quality segmentations across all datasets. Rigorous validation, including expert physician evaluation and automatic quality control, was conducted to validate the resulting masks. Additionally, we provide individualized quality indices per mask and an overall quality estimation per dataset. This dataset serves as a valuable resource for the broader scientific community, streamlining the development and assessment of innovative methodologies in chest X-ray analysis. The CheXmask dataset is publicly available at: \url{https://physionet.org/content/chexmask-cxr-segmentation-data/}.
Towards unraveling calibration biases in medical image analysis
May 09, 2023


Abstract:In recent years the development of artificial intelligence (AI) systems for automated medical image analysis has gained enormous momentum. At the same time, a large body of work has shown that AI systems can systematically and unfairly discriminate against certain populations in various application scenarios. These two facts have motivated the emergence of algorithmic fairness studies in this field. Most research on healthcare algorithmic fairness to date has focused on the assessment of biases in terms of classical discrimination metrics such as AUC and accuracy. Potential biases in terms of model calibration, however, have only recently begun to be evaluated. This is especially important when working with clinical decision support systems, as predictive uncertainty is key for health professionals to optimally evaluate and combine multiple sources of information. In this work we study discrimination and calibration biases in models trained for automatic detection of malignant dermatological conditions from skin lesions images. Importantly, we show how several typically employed calibration metrics are systematically biased with respect to sample sizes, and how this can lead to erroneous fairness analysis if not taken into consideration. This is of particular relevance to fairness studies, where data imbalance results in drastic sample size differences between demographic sub-groups, which, if not taken into account, can act as confounders.
Improving anatomical plausibility in medical image segmentation via hybrid graph neural networks: applications to chest x-ray analysis
Apr 01, 2022

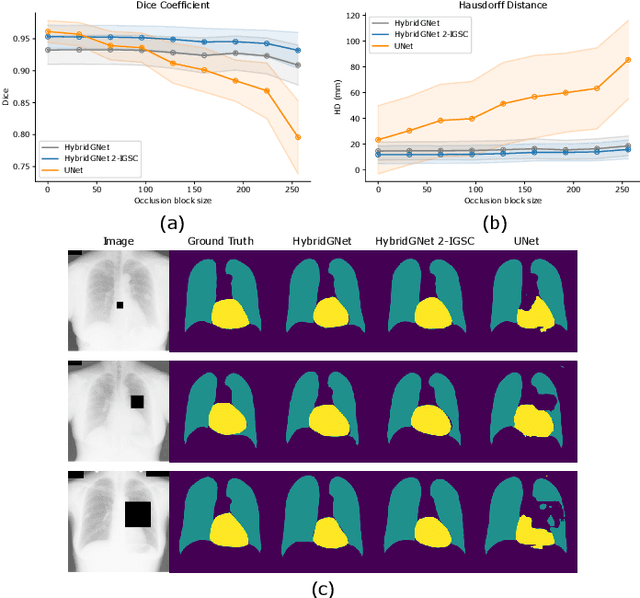

Abstract:Anatomical segmentation is a fundamental task in medical image computing, generally tackled with fully convolutional neural networks which produce dense segmentation masks. These models are often trained with loss functions such as cross-entropy or Dice, which assume pixels to be independent of each other, thus ignoring topological errors and anatomical inconsistencies. We address this limitation by moving from pixel-level to graph representations, which allow to naturally incorporate anatomical constraints by construction. To this end, we introduce HybridGNet, an encoder-decoder neural architecture that leverages standard convolutions for image feature encoding and graph convolutional neural networks (GCNNs) to decode plausible representations of anatomical structures. We also propose a novel image-to-graph skip connection layer which allows localized features to flow from standard convolutional blocks to GCNN blocks, and show that it improves segmentation accuracy. The proposed architecture is extensively evaluated in a variety of domain shift and image occlusion scenarios, and audited considering different types of demographic domain shift. Our comprehensive experimental setup compares HybridGNet with other landmark and pixel-based models for anatomical segmentation in chest x-ray images, and shows that it produces anatomically plausible results in challenging scenarios where other models tend to fail.
Understanding the impact of class imbalance on the performance of chest x-ray image classifiers
Dec 23, 2021



Abstract:This work aims to understand the impact of class imbalance on the performance of chest x-ray classifiers, in light of the standard evaluation practices adopted by researchers in terms of discrimination and calibration performance. Firstly, we conducted a literature study to analyze common scientific practices and confirmed that: (1) even when dealing with highly imbalanced datasets, the community tends to use metrics that are dominated by the majority class; and (2) it is still uncommon to include calibration studies for chest x-ray classifiers, albeit its importance in the context of healthcare. Secondly, we perform a systematic experiment on two major chest x-ray datasets to explore the behavior of several performance metrics under different class ratios and show that widely adopted metrics can conceal the performance in the minority class. Finally, we propose the adoption of two alternative metrics, the precision-recall curve and the Balanced Brier score, which better reflect the performance of the system in such scenarios. Our results indicate that current evaluation practices adopted by the research community for chest x-ray classifiers may not reflect the performance of such systems for computer-aided diagnosis in real clinical scenarios, and suggest alternatives to improve this situation.
Chest x-ray automated triage: a semiologic approach designed for clinical implementation, exploiting different types of labels through a combination of four Deep Learning architectures
Dec 23, 2020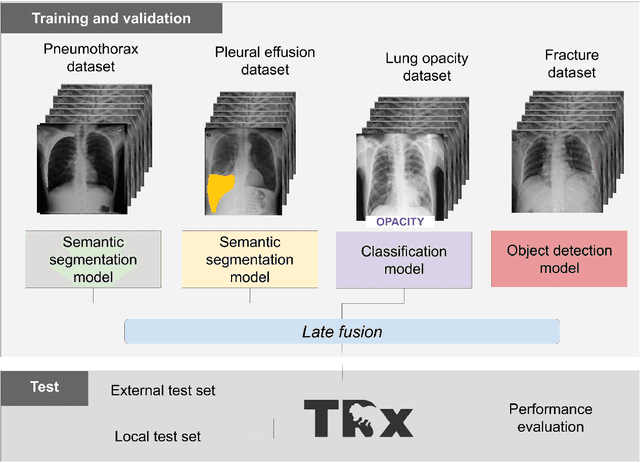
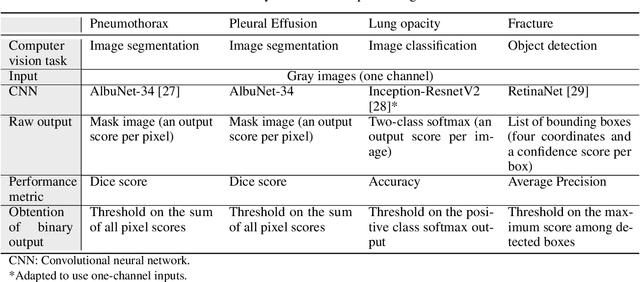
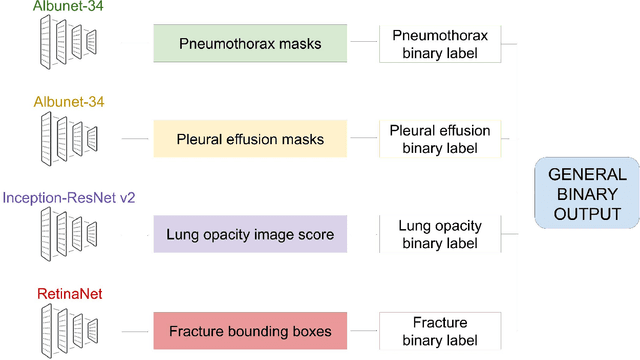
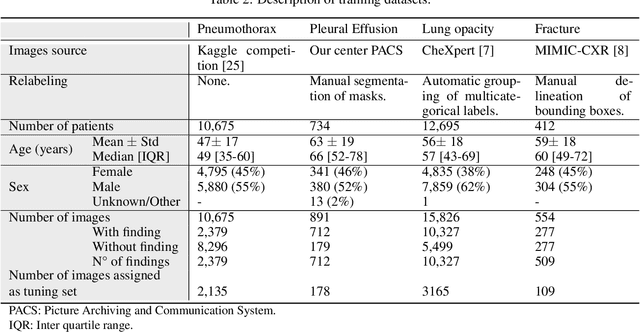
Abstract:BACKGROUND AND OBJECTIVES: The multiple chest x-ray datasets released in the last years have ground-truth labels intended for different computer vision tasks, suggesting that performance in automated chest-xray interpretation might improve by using a method that can exploit diverse types of annotations. This work presents a Deep Learning method based on the late fusion of different convolutional architectures, that allows training with heterogeneous data with a simple implementation, and evaluates its performance on independent test data. We focused on obtaining a clinically useful tool that could be successfully integrated into a hospital workflow. MATERIALS AND METHODS: Based on expert opinion, we selected four target chest x-ray findings, namely lung opacities, fractures, pneumothorax and pleural effusion. For each finding we defined the most adequate type of ground-truth label, and built four training datasets combining images from public chest x-ray datasets and our institutional archive. We trained four different Deep Learning architectures and combined their outputs with a late fusion strategy, obtaining a unified tool. Performance was measured on two test datasets: an external openly-available dataset, and a retrospective institutional dataset, to estimate performance on local population. RESULTS: The external and local test sets had 4376 and 1064 images, respectively, for which the model showed an area under the Receiver Operating Characteristics curve of 0.75 (95%CI: 0.74-0.76) and 0.87 (95%CI: 0.86-0.89) in the detection of abnormal chest x-rays. For the local population, a sensitivity of 86% (95%CI: 84-90), and a specificity of 88% (95%CI: 86-90) were obtained, with no significant differences between demographic subgroups. We present examples of heatmaps to show the accomplished level of interpretability, examining true and false positives.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge