Shahabedin Nabavi
Faculty of Computer Science and Engineering, Shahid Beheshti University, Tehran, Iran
FractMorph: A Fractional Fourier-Based Multi-Domain Transformer for Deformable Image Registration
Aug 17, 2025Abstract:Deformable image registration (DIR) is a crucial and challenging technique for aligning anatomical structures in medical images and is widely applied in diverse clinical applications. However, existing approaches often struggle to capture fine-grained local deformations and large-scale global deformations simultaneously within a unified framework. We present FractMorph, a novel 3D dual-parallel transformer-based architecture that enhances cross-image feature matching through multi-domain fractional Fourier transform (FrFT) branches. Each Fractional Cross-Attention (FCA) block applies parallel FrFTs at fractional angles of 0{\deg}, 45{\deg}, 90{\deg}, along with a log-magnitude branch, to effectively extract local, semi-global, and global features at the same time. These features are fused via cross-attention between the fixed and moving image streams. A lightweight U-Net style network then predicts a dense deformation field from the transformer-enriched features. On the ACDC cardiac MRI dataset, FractMorph achieves state-of-the-art performance with an overall Dice Similarity Coefficient (DSC) of 86.45%, an average per-structure DSC of 75.15%, and a 95th-percentile Hausdorff distance (HD95) of 1.54 mm on our data split. We also introduce FractMorph-Light, a lightweight variant of our model with only 29.6M parameters, which maintains the superior accuracy of the main model while using approximately half the memory. Our results demonstrate that multi-domain spectral-spatial attention in transformers can robustly and efficiently model complex non-rigid deformations in medical images using a single end-to-end network, without the need for scenario-specific tuning or hierarchical multi-scale networks. The source code of our implementation is available at https://github.com/shayankebriti/FractMorph.
Small Object Detection: A Comprehensive Survey on Challenges, Techniques and Real-World Applications
Mar 26, 2025Abstract:Small object detection (SOD) is a critical yet challenging task in computer vision, with applications like spanning surveillance, autonomous systems, medical imaging, and remote sensing. Unlike larger objects, small objects contain limited spatial and contextual information, making accurate detection difficult. Challenges such as low resolution, occlusion, background interference, and class imbalance further complicate the problem. This survey provides a comprehensive review of recent advancements in SOD using deep learning, focusing on articles published in Q1 journals during 2024-2025. We analyzed challenges, state-of-the-art techniques, datasets, evaluation metrics, and real-world applications. Recent advancements in deep learning have introduced innovative solutions, including multi-scale feature extraction, Super-Resolution (SR) techniques, attention mechanisms, and transformer-based architectures. Additionally, improvements in data augmentation, synthetic data generation, and transfer learning have addressed data scarcity and domain adaptation issues. Furthermore, emerging trends such as lightweight neural networks, knowledge distillation (KD), and self-supervised learning offer promising directions for improving detection efficiency, particularly in resource-constrained environments like Unmanned Aerial Vehicles (UAV)-based surveillance and edge computing. We also review widely used datasets, along with standard evaluation metrics such as mean Average Precision (mAP) and size-specific AP scores. The survey highlights real-world applications, including traffic monitoring, maritime surveillance, industrial defect detection, and precision agriculture. Finally, we discuss open research challenges and future directions, emphasizing the need for robust domain adaptation techniques, better feature fusion strategies, and real-time performance optimization.
Towards Universal Learning-based Model for Cardiac Image Reconstruction: Summary of the CMRxRecon2024 Challenge
Mar 05, 2025Abstract:Cardiovascular magnetic resonance (CMR) offers diverse imaging contrasts for assessment of cardiac function and tissue characterization. However, acquiring each single CMR modality is often time-consuming, and comprehensive clinical protocols require multiple modalities with various sampling patterns, further extending the overall acquisition time and increasing susceptibility to motion artifacts. Existing deep learning-based reconstruction methods are often designed for specific acquisition parameters, which limits their ability to generalize across a variety of scan scenarios. As part of the CMRxRecon Series, the CMRxRecon2024 challenge provides diverse datasets encompassing multi-modality multi-view imaging with various sampling patterns, and a platform for the international community to develop and benchmark reconstruction solutions in two well-crafted tasks. Task 1 is a modality-universal setting, evaluating the out-of-distribution generalization of the reconstructed model, while Task 2 follows sampling-universal setting assessing the one-for-all adaptability of the universal model. Main contributions include providing the first and largest publicly available multi-modality, multi-view cardiac k-space dataset; developing a benchmarking platform that simulates clinical acceleration protocols, with a shared code library and tutorial for various k-t undersampling patterns and data processing; giving technical insights of enhanced data consistency based on physic-informed networks and adaptive prompt-learning embedding to be versatile to different clinical settings; additional finding on evaluation metrics to address the limitations of conventional ground-truth references in universal reconstruction tasks.
An All-in-one Approach for Accelerated Cardiac MRI Reconstruction
Nov 16, 2024Abstract:Cardiovascular magnetic resonance (CMR) imaging is the gold standard for diagnosing several heart diseases due to its non-invasive nature and proper contrast. MR imaging is time-consuming because of signal acquisition and image formation issues. Prolonging the imaging process can result in the appearance of artefacts in the final image, which can affect the diagnosis. It is possible to speed up CMR imaging using image reconstruction based on deep learning. For this purpose, the high-quality clinical interpretable images can be reconstructed by acquiring highly undersampled k-space data, that is only partially filled, and using a deep learning model. In this study, we proposed a stepwise reconstruction approach based on the Patch-GAN structure for highly undersampled k-space data compatible with the multi-contrast nature, various anatomical views and trajectories of CMR imaging. The proposed approach was validated using the CMRxRecon2024 challenge dataset and outperformed previous studies. The structural similarity index measure (SSIM) values for the first and second tasks of the challenge are 99.07 and 97.99, respectively. This approach can accelerate CMR imaging to obtain high-quality images, more accurate diagnosis and a pleasant patient experience.
Statistical Distance-Guided Unsupervised Domain Adaptation for Automated Multi-Class Cardiovascular Magnetic Resonance Image Quality Assessment
Aug 31, 2024Abstract:This study proposes an attention-based statistical distance-guided unsupervised domain adaptation model for multi-class cardiovascular magnetic resonance (CMR) image quality assessment. The proposed model consists of a feature extractor, a label predictor and a statistical distance estimator. An annotated dataset as the source set and an unlabeled dataset as the target set with different statistical distributions are considered inputs. The statistical distance estimator approximates the Wasserstein distance between the extracted feature vectors from the source and target data in a mini-batch. The label predictor predicts data labels of source data and uses a combinational loss function for training, which includes cross entropy and centre loss functions plus the estimated value of the distance estimator. Four datasets, including imaging and k-space data, were used to evaluate the proposed model in identifying four common CMR imaging artefacts: respiratory and cardiac motions, Gibbs ringing and Aliasing. The results of the extensive experiments showed that the proposed model, both in image and k-space analysis, has an acceptable performance in covering the domain shift between the source and target sets. The model explainability evaluations and the ablation studies confirmed the proper functioning and effectiveness of all the model's modules. The proposed model outperformed the previous studies regarding performance and the number of examined artefacts. The proposed model can be used for CMR post-imaging quality control or in large-scale cohort studies for image and k-space quality assessment due to the appropriate performance in domain shift coverage without a tedious data-labelling process.
Multiple Teachers-Meticulous Student: A Domain Adaptive Meta-Knowledge Distillation Model for Medical Image Classification
Mar 17, 2024



Abstract:Background: Image classification can be considered one of the key pillars of medical image analysis. Deep learning (DL) faces challenges that prevent its practical applications despite the remarkable improvement in medical image classification. The data distribution differences can lead to a drop in the efficiency of DL, known as the domain shift problem. Besides, requiring bulk annotated data for model training, the large size of models, and the privacy-preserving of patients are other challenges of using DL in medical image classification. This study presents a strategy that can address the mentioned issues simultaneously. Method: The proposed domain adaptive model based on knowledge distillation can classify images by receiving limited annotated data of different distributions. The designed multiple teachers-meticulous student model trains a student network that tries to solve the challenges by receiving the parameters of several teacher networks. The proposed model was evaluated using six available datasets of different distributions by defining the respiratory motion artefact detection task. Results: The results of extensive experiments using several datasets show the superiority of the proposed model in addressing the domain shift problem and lack of access to bulk annotated data. Besides, the privacy preservation of patients by receiving only the teacher network parameters instead of the original data and consolidating the knowledge of several DL models into a model with almost similar performance are other advantages of the proposed model. Conclusions: The proposed model can pave the way for practical clinical applications of deep classification methods by achieving the mentioned objectives simultaneously.
A Cascade Transformer-based Model for 3D Dose Distribution Prediction in Head and Neck Cancer Radiotherapy
Jul 22, 2023Abstract:Radiation therapy is the primary method used to treat cancer in the clinic. Its goal is to deliver a precise dose to the planning target volume (PTV) while protecting the surrounding organs at risk (OARs). However, the traditional workflow used by dosimetrists to plan the treatment is time-consuming and subjective, requiring iterative adjustments based on their experience. Deep learning methods can be used to predict dose distribution maps to address these limitations. The study proposes a cascade model for organs at risk segmentation and dose distribution prediction. An encoder-decoder network has been developed for the segmentation task, in which the encoder consists of transformer blocks, and the decoder uses multi-scale convolutional blocks. Another cascade encoder-decoder network has been proposed for dose distribution prediction using a pyramid architecture. The proposed model has been evaluated using an in-house head and neck cancer dataset of 96 patients and OpenKBP, a public head and neck cancer dataset of 340 patients. The segmentation subnet achieved 0.79 and 2.71 for Dice and HD95 scores, respectively. This subnet outperformed the existing baselines. The dose distribution prediction subnet outperformed the winner of the OpenKBP2020 competition with 2.77 and 1.79 for dose and DVH scores, respectively. The predicted dose maps showed good coincidence with ground truth, with a superiority after linking with the auxiliary segmentation task. The proposed model outperformed state-of-the-art methods, especially in regions with low prescribed doses.
A Generalised Deep Meta-Learning Model for Automated Quality Control of Cardiovascular Magnetic Resonance Images
Mar 23, 2023


Abstract:Background and Objectives: Cardiovascular magnetic resonance (CMR) imaging is a powerful modality in functional and anatomical assessment for various cardiovascular diseases. Sufficient image quality is essential to achieve proper diagnosis and treatment. A large number of medical images, the variety of imaging artefacts, and the workload of imaging centres are among the things that reveal the necessity of automatic image quality assessment (IQA). However, automated IQA requires access to bulk annotated datasets for training deep learning (DL) models. Labelling medical images is a tedious, costly and time-consuming process, which creates a fundamental challenge in proposing DL-based methods for medical applications. This study aims to present a new method for CMR IQA when there is limited access to annotated datasets. Methods: The proposed generalised deep meta-learning model can evaluate the quality by learning tasks in the prior stage and then fine-tuning the resulting model on a small labelled dataset of the desired tasks. This model was evaluated on the data of over 6,000 subjects from the UK Biobank for five defined tasks, including detecting respiratory motion, cardiac motion, Aliasing and Gibbs ringing artefacts and images without artefacts. Results: The results of extensive experiments show the superiority of the proposed model. Besides, comparing the model's accuracy with the domain adaptation model indicates a significant difference by using only 64 annotated images related to the desired tasks. Conclusion: The proposed model can identify unknown artefacts in images with acceptable accuracy, which makes it suitable for medical applications and quality assessment of large cohorts.
Stacked Cross-modal Feature Consolidation Attention Networks for Image Captioning
Feb 08, 2023Abstract:Recently, the attention-enriched encoder-decoder framework has aroused great interest in image captioning due to its overwhelming progress. Many visual attention models directly leverage meaningful regions to generate image descriptions. However, seeking a direct transition from visual space to text is not enough to generate fine-grained captions. This paper exploits a feature-compounding approach to bring together high-level semantic concepts and visual information regarding the contextual environment fully end-to-end. Thus, we propose a stacked cross-modal feature consolidation (SCFC) attention network for image captioning in which we simultaneously consolidate cross-modal features through a novel compounding function in a multi-step reasoning fashion. Besides, we jointly employ spatial information and context-aware attributes (CAA) as the principal components in our proposed compounding function, where our CAA provides a concise context-sensitive semantic representation. To make better use of consolidated features potential, we further propose an SCFC-LSTM as the caption generator, which can leverage discriminative semantic information through the caption generation process. The experimental results indicate that our proposed SCFC can outperform various state-of-the-art image captioning benchmarks in terms of popular metrics on the MSCOCO and Flickr30K datasets.
Fully Automated Assessment of Cardiac Coverage in Cine Cardiovascular Magnetic Resonance Images using an Explainable Deep Visual Salient Region Detection Model
Jun 14, 2022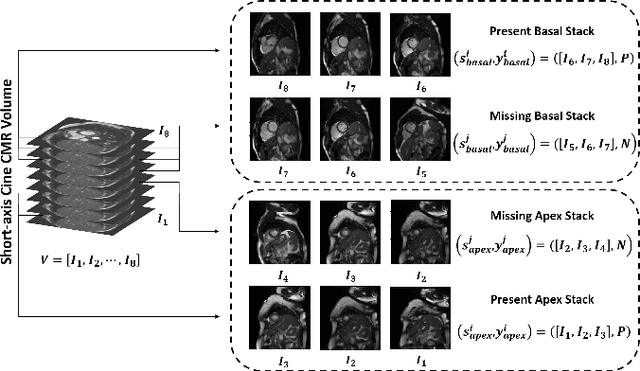
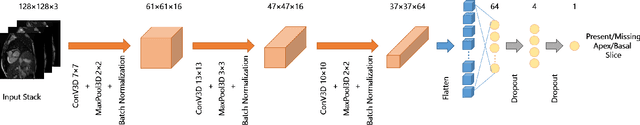
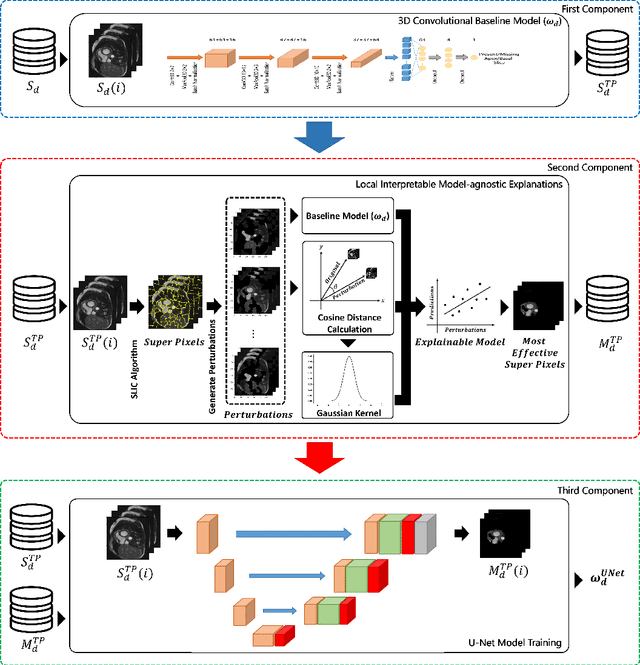
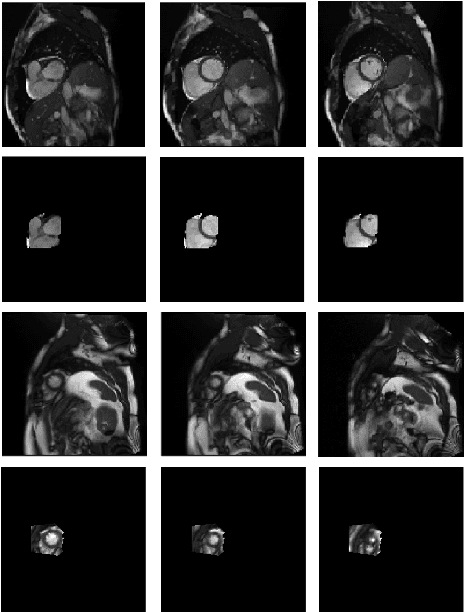
Abstract:Cardiovascular magnetic resonance (CMR) imaging has become a modality with superior power for the diagnosis and prognosis of cardiovascular diseases. One of the essential basic quality controls of CMR images is to investigate the complete cardiac coverage, which is necessary for the volumetric and functional assessment. This study examines the full cardiac coverage using a 3D convolutional model and then reduces the number of false predictions using an innovative salient region detection model. Salient regions are extracted from the short-axis cine CMR stacks using a three-step proposed algorithm. Combining the 3D CNN baseline model with the proposed salient region detection model provides a cascade detector that can reduce the number of false negatives of the baseline model. The results obtained on the images of over 6,200 participants of the UK Biobank population cohort study show the superiority of the proposed model over the previous state-of-the-art studies. The dataset is the largest regarding the number of participants to control the cardiac coverage. The accuracy of the baseline model in identifying the presence/absence of basal/apical slices is 96.25\% and 94.51\%, respectively, which increases to 96.88\% and 95.72\% after improving using the proposed salient region detection model. Using the salient region detection model by forcing the baseline model to focus on the most informative areas of the images can help the model correct misclassified samples' predictions. The proposed fully automated model's performance indicates that this model can be used in image quality control in population cohort datasets and also real-time post-imaging quality assessments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge