Wenqi Lu
SACB-Net: Spatial-awareness Convolutions for Medical Image Registration
Mar 25, 2025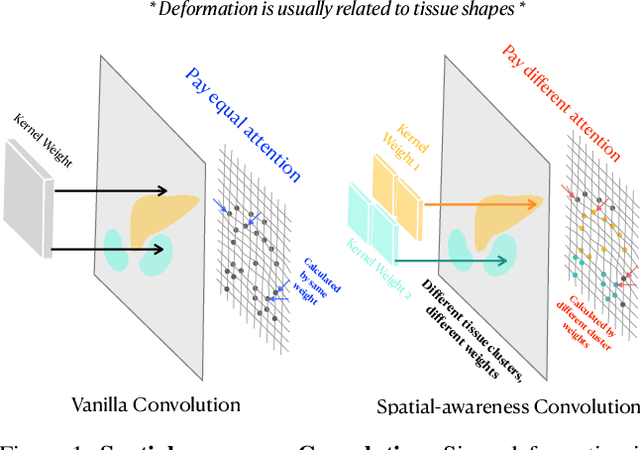
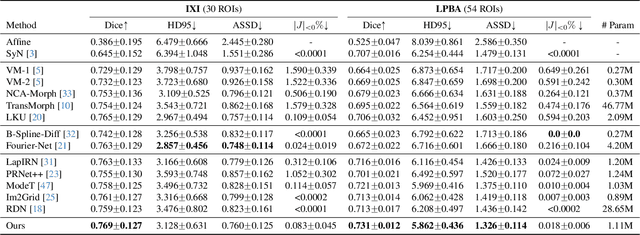
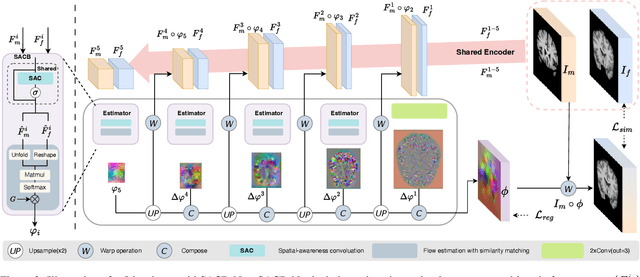
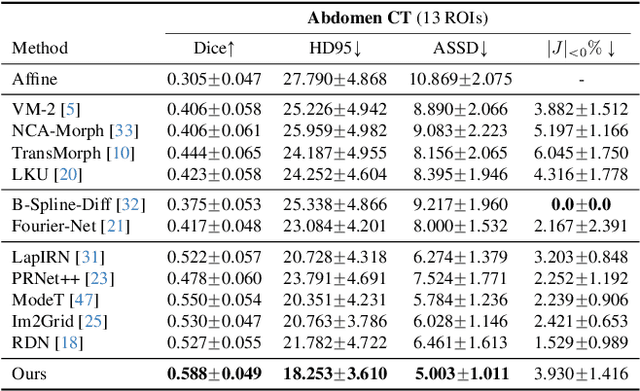
Abstract:Deep learning-based image registration methods have shown state-of-the-art performance and rapid inference speeds. Despite these advances, many existing approaches fall short in capturing spatially varying information in non-local regions of feature maps due to the reliance on spatially-shared convolution kernels. This limitation leads to suboptimal estimation of deformation fields. In this paper, we propose a 3D Spatial-Awareness Convolution Block (SACB) to enhance the spatial information within feature representations. Our SACB estimates the spatial clusters within feature maps by leveraging feature similarity and subsequently parameterizes the adaptive convolution kernels across diverse regions. This adaptive mechanism generates the convolution kernels (weights and biases) tailored to spatial variations, thereby enabling the network to effectively capture spatially varying information. Building on SACB, we introduce a pyramid flow estimator (named SACB-Net) that integrates SACBs to facilitate multi-scale flow composition, particularly addressing large deformations. Experimental results on the brain IXI and LPBA datasets as well as Abdomen CT datasets demonstrate the effectiveness of SACB and the superiority of SACB-Net over the state-of-the-art learning-based registration methods. The code is available at https://github.com/x-xc/SACB_Net .
WiNet: Wavelet-based Incremental Learning for Efficient Medical Image Registration
Jul 18, 2024Abstract:Deep image registration has demonstrated exceptional accuracy and fast inference. Recent advances have adopted either multiple cascades or pyramid architectures to estimate dense deformation fields in a coarse-to-fine manner. However, due to the cascaded nature and repeated composition/warping operations on feature maps, these methods negatively increase memory usage during training and testing. Moreover, such approaches lack explicit constraints on the learning process of small deformations at different scales, thus lacking explainability. In this study, we introduce a model-driven WiNet that incrementally estimates scale-wise wavelet coefficients for the displacement/velocity field across various scales, utilizing the wavelet coefficients derived from the original input image pair. By exploiting the properties of the wavelet transform, these estimated coefficients facilitate the seamless reconstruction of a full-resolution displacement/velocity field via our devised inverse discrete wavelet transform (IDWT) layer. This approach avoids the complexities of cascading networks or composition operations, making our WiNet an explainable and efficient competitor with other coarse-to-fine methods. Extensive experimental results from two 3D datasets show that our WiNet is accurate and GPU efficient. The code is available at https://github.com/x-xc/WiNet .
Development of Automated Neural Network Prediction for Echocardiographic Left ventricular Ejection Fraction
Mar 18, 2024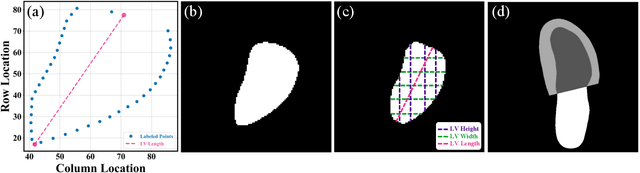

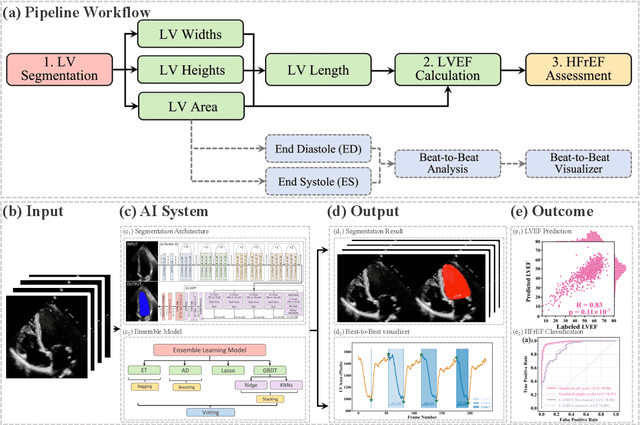
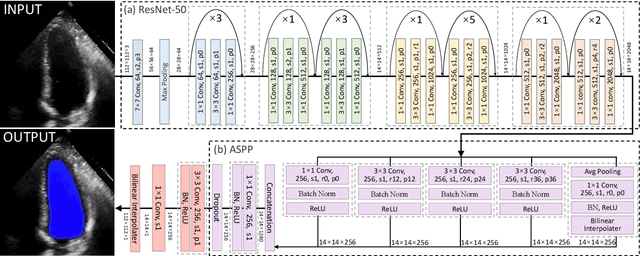
Abstract:The echocardiographic measurement of left ventricular ejection fraction (LVEF) is fundamental to the diagnosis and classification of patients with heart failure (HF). In order to quantify LVEF automatically and accurately, this paper proposes a new pipeline method based on deep neural networks and ensemble learning. Within the pipeline, an Atrous Convolutional Neural Network (ACNN) was first trained to segment the left ventricle (LV), before employing the area-length formulation based on the ellipsoid single-plane model to calculate LVEF values. This formulation required inputs of LV area, derived from segmentation using an improved Jeffrey's method, as well as LV length, derived from a novel ensemble learning model. To further improve the pipeline's accuracy, an automated peak detection algorithm was used to identify end-diastolic and end-systolic frames, avoiding issues with human error. Subsequently, single-beat LVEF values were averaged across all cardiac cycles to obtain the final LVEF. This method was developed and internally validated in an open-source dataset containing 10,030 echocardiograms. The Pearson's correlation coefficient was 0.83 for LVEF prediction compared to expert human analysis (p<0.001), with a subsequent area under the receiver operator curve (AUROC) of 0.98 (95% confidence interval 0.97 to 0.99) for categorisation of HF with reduced ejection (HFrEF; LVEF<40%). In an external dataset with 200 echocardiograms, this method achieved an AUC of 0.90 (95% confidence interval 0.88 to 0.91) for HFrEF assessment. This study demonstrates that an automated neural network-based calculation of LVEF is comparable to expert clinicians performing time-consuming, frame-by-frame manual evaluation of cardiac systolic function.
Decoder-Only Image Registration
Feb 05, 2024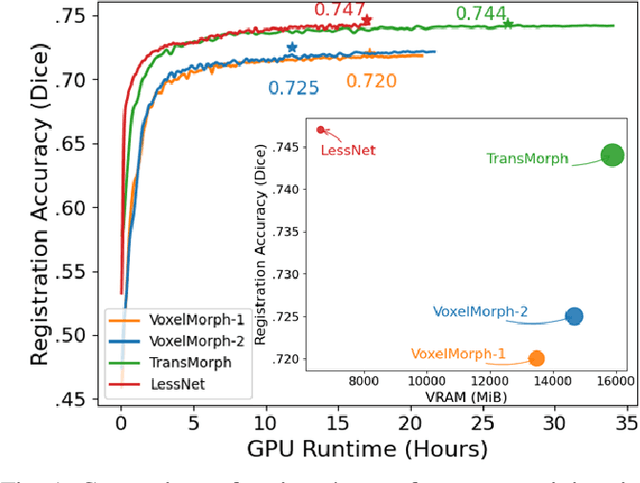
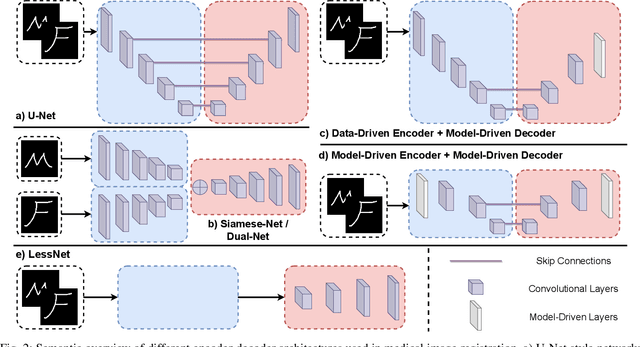
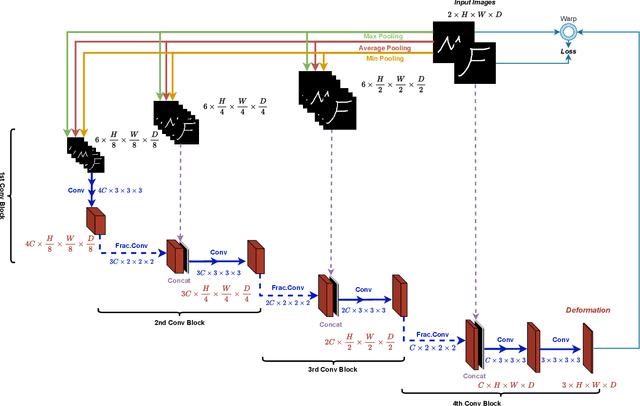
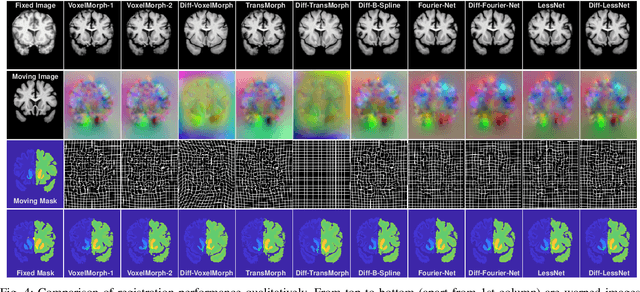
Abstract:In unsupervised medical image registration, the predominant approaches involve the utilization of a encoder-decoder network architecture, allowing for precise prediction of dense, full-resolution displacement fields from given paired images. Despite its widespread use in the literature, we argue for the necessity of making both the encoder and decoder learnable in such an architecture. For this, we propose a novel network architecture, termed LessNet in this paper, which contains only a learnable decoder, while entirely omitting the utilization of a learnable encoder. LessNet substitutes the learnable encoder with simple, handcrafted features, eliminating the need to learn (optimize) network parameters in the encoder altogether. Consequently, this leads to a compact, efficient, and decoder-only architecture for 3D medical image registration. Evaluated on two publicly available brain MRI datasets, we demonstrate that our decoder-only LessNet can effectively and efficiently learn both dense displacement and diffeomorphic deformation fields in 3D. Furthermore, our decoder-only LessNet can achieve comparable registration performance to state-of-the-art methods such as VoxelMorph and TransMorph, while requiring significantly fewer computational resources. Our code and pre-trained models are available at https://github.com/xi-jia/LessNet.
Optimizing ADMM and Over-Relaxed ADMM Parameters for Linear Quadratic Problems
Jan 01, 2024



Abstract:The Alternating Direction Method of Multipliers (ADMM) has gained significant attention across a broad spectrum of machine learning applications. Incorporating the over-relaxation technique shows potential for enhancing the convergence rate of ADMM. However, determining optimal algorithmic parameters, including both the associated penalty and relaxation parameters, often relies on empirical approaches tailored to specific problem domains and contextual scenarios. Incorrect parameter selection can significantly hinder ADMM's convergence rate. To address this challenge, in this paper we first propose a general approach to optimize the value of penalty parameter, followed by a novel closed-form formula to compute the optimal relaxation parameter in the context of linear quadratic problems (LQPs). We then experimentally validate our parameter selection methods through random instantiations and diverse imaging applications, encompassing diffeomorphic image registration, image deblurring, and MRI reconstruction.
Fourier-Net+: Leveraging Band-Limited Representation for Efficient 3D Medical Image Registration
Jul 06, 2023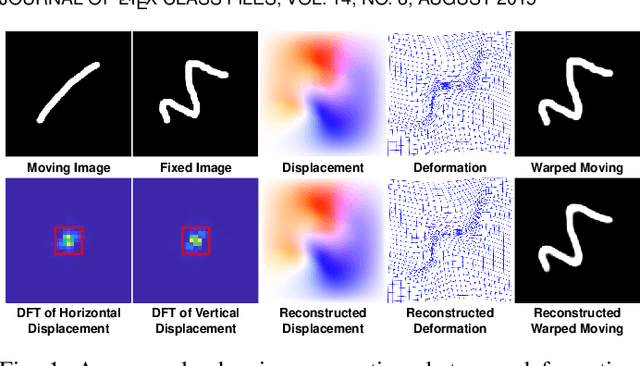
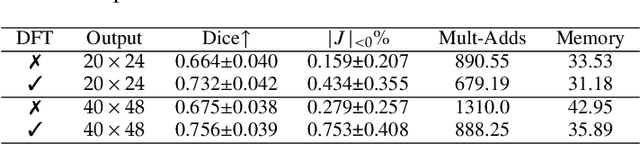
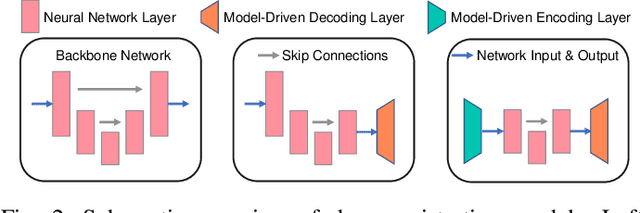
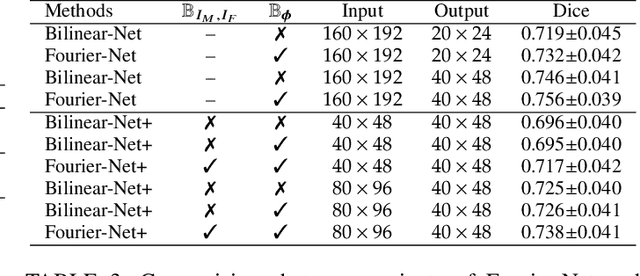
Abstract:U-Net style networks are commonly utilized in unsupervised image registration to predict dense displacement fields, which for high-resolution volumetric image data is a resource-intensive and time-consuming task. To tackle this challenge, we first propose Fourier-Net, which replaces the costly U-Net style expansive path with a parameter-free model-driven decoder. Instead of directly predicting a full-resolution displacement field, our Fourier-Net learns a low-dimensional representation of the displacement field in the band-limited Fourier domain which our model-driven decoder converts to a full-resolution displacement field in the spatial domain. Expanding upon Fourier-Net, we then introduce Fourier-Net+, which additionally takes the band-limited spatial representation of the images as input and further reduces the number of convolutional layers in the U-Net style network's contracting path. Finally, to enhance the registration performance, we propose a cascaded version of Fourier-Net+. We evaluate our proposed methods on three datasets, on which our proposed Fourier-Net and its variants achieve comparable results with current state-of-the art methods, while exhibiting faster inference speeds, lower memory footprint, and fewer multiply-add operations. With such small computational cost, our Fourier-Net+ enables the efficient training of large-scale 3D registration on low-VRAM GPUs. Our code is publicly available at \url{https://github.com/xi-jia/Fourier-Net}.
Fourier-Net: Fast Image Registration with Band-limited Deformation
Nov 29, 2022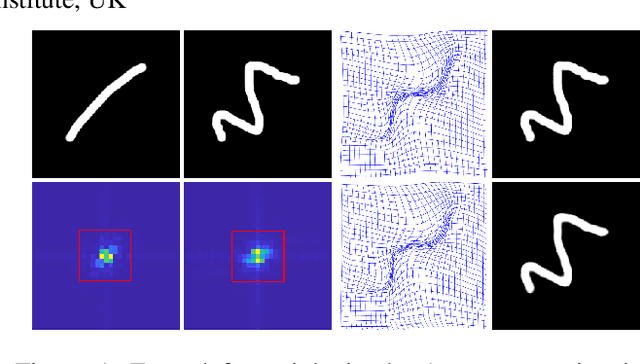
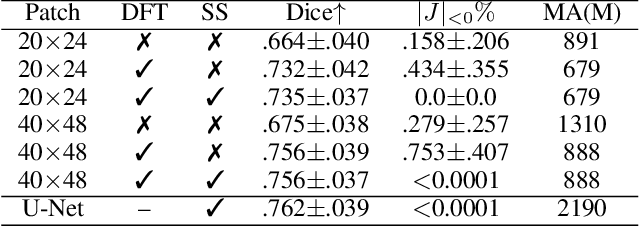
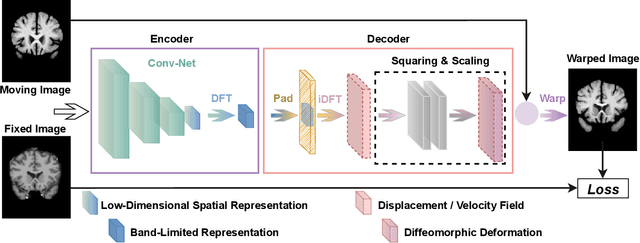
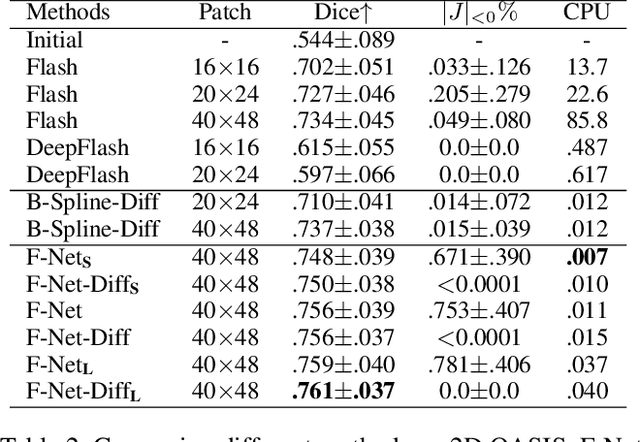
Abstract:Unsupervised image registration commonly adopts U-Net style networks to predict dense displacement fields in the full-resolution spatial domain. For high-resolution volumetric image data, this process is however resource intensive and time-consuming. To tackle this problem, we propose the Fourier-Net, replacing the expansive path in a U-Net style network with a parameter-free model-driven decoder. Specifically, instead of our Fourier-Net learning to output a full-resolution displacement field in the spatial domain, we learn its low-dimensional representation in a band-limited Fourier domain. This representation is then decoded by our devised model-driven decoder (consisting of a zero padding layer and an inverse discrete Fourier transform layer) to the dense, full-resolution displacement field in the spatial domain. These changes allow our unsupervised Fourier-Net to contain fewer parameters and computational operations, resulting in faster inference speeds. Fourier-Net is then evaluated on two public 3D brain datasets against various state-of-the-art approaches. For example, when compared to a recent transformer-based method, i.e., TransMorph, our Fourier-Net, only using 0.22$\%$ of its parameters and 6.66$\%$ of the mult-adds, achieves a 0.6\% higher Dice score and an 11.48$\times$ faster inference speed. Code is available at \url{https://github.com/xi-jia/Fourier-Net}.
U-Net vs Transformer: Is U-Net Outdated in Medical Image Registration?
Aug 13, 2022
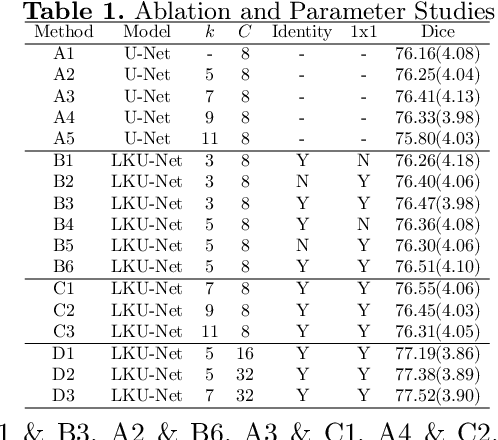
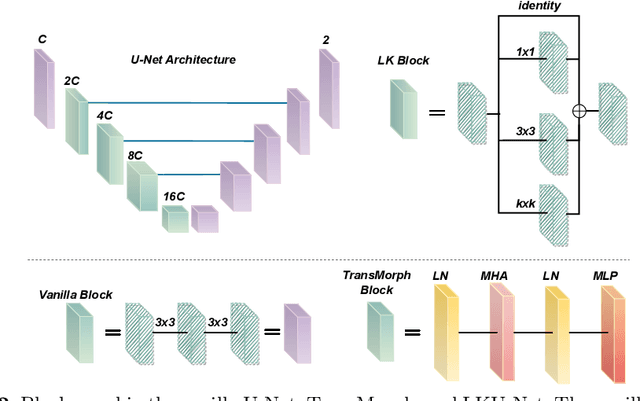
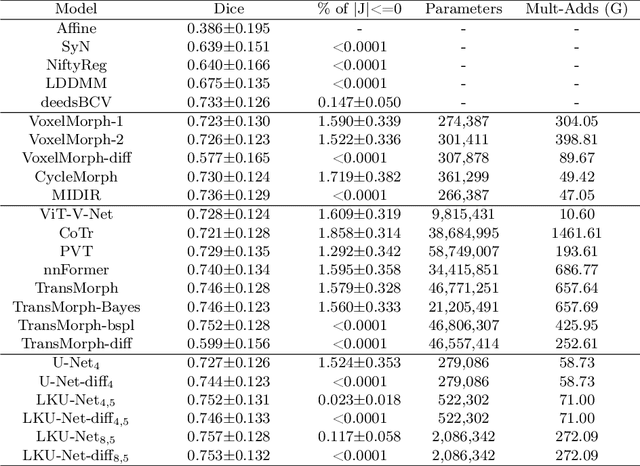
Abstract:Due to their extreme long-range modeling capability, vision transformer-based networks have become increasingly popular in deformable image registration. We believe, however, that the receptive field of a 5-layer convolutional U-Net is sufficient to capture accurate deformations without needing long-range dependencies. The purpose of this study is therefore to investigate whether U-Net-based methods are outdated compared to modern transformer-based approaches when applied to medical image registration. For this, we propose a large kernel U-Net (LKU-Net) by embedding a parallel convolutional block to a vanilla U-Net in order to enhance the effective receptive field. On the public 3D IXI brain dataset for atlas-based registration, we show that the performance of the vanilla U-Net is already comparable with that of state-of-the-art transformer-based networks (such as TransMorph), and that the proposed LKU-Net outperforms TransMorph by using only 1.12% of its parameters and 10.8% of its mult-adds operations. We further evaluate LKU-Net on a MICCAI Learn2Reg 2021 challenge dataset for inter-subject registration, our LKU-Net also outperforms TransMorph on this dataset and ranks first on the public leaderboard as of the submission of this work. With only modest modifications to the vanilla U-Net, we show that U-Net can outperform transformer-based architectures on inter-subject and atlas-based 3D medical image registration. Code is available at https://github.com/xi-jia/LKU-Net.
SlideGraph+: Whole Slide Image Level Graphs to Predict HER2Status in Breast Cancer
Oct 12, 2021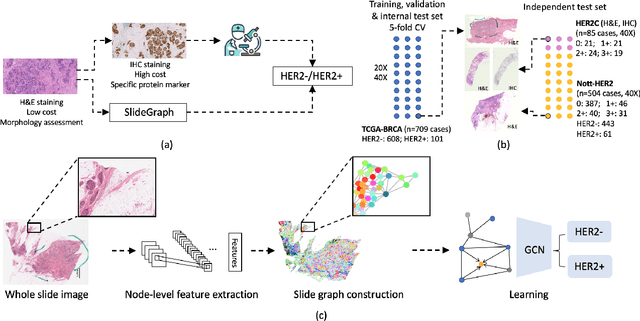
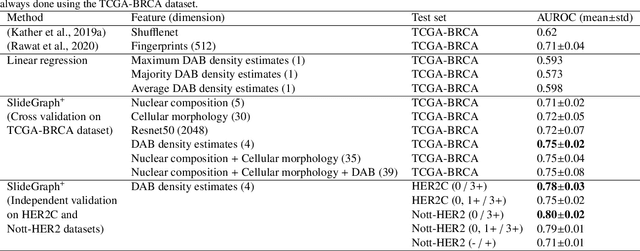
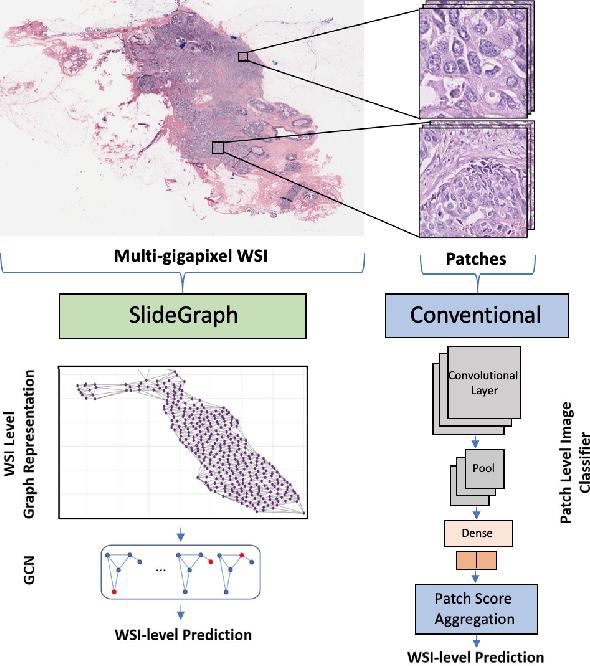
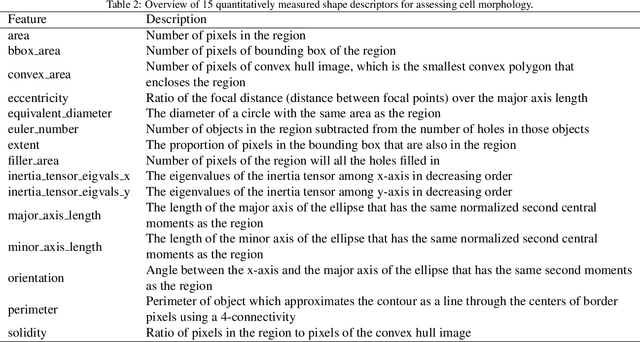
Abstract:Human epidermal growth factor receptor 2 (HER2) is an important prognostic and predictive factor which is overexpressed in 15-20% of breast cancer (BCa). The determination of its status is a key clinical decision making step for selection of treatment regimen and prognostication. HER2 status is evaluated using transcroptomics or immunohistochemistry (IHC) through situ hybridisation (ISH) which require additional costs and tissue burden in addition to analytical variabilities in terms of manual observational biases in scoring. In this study, we propose a novel graph neural network (GNN) based model (termed SlideGraph+) to predict HER2 status directly from whole-slide images of routine Haematoxylin and Eosin (H&E) slides. The network was trained and tested on slides from The Cancer Genome Atlas (TCGA) in addition to two independent test datasets. We demonstrate that the proposed model outperforms the state-of-the-art methods with area under the ROC curve (AUC) values > 0.75 on TCGA and 0.8 on independent test sets. Our experiments show that the proposed approach can be utilised for case triaging as well as pre-ordering diagnostic tests in a diagnostic setting. It can also be used for other weakly supervised prediction problems in computational pathology. The SlideGraph+ code is available at https://github.com/wenqi006/SlideGraph.
Semantic annotation for computational pathology: Multidisciplinary experience and best practice recommendations
Jun 25, 2021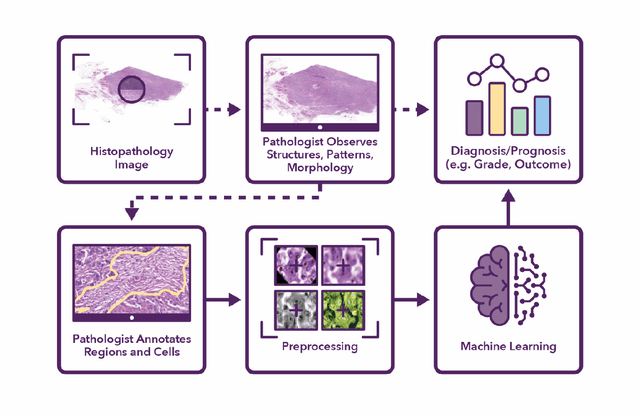
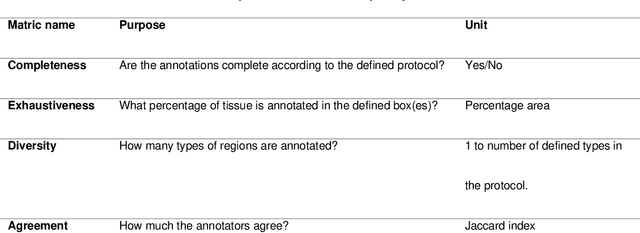
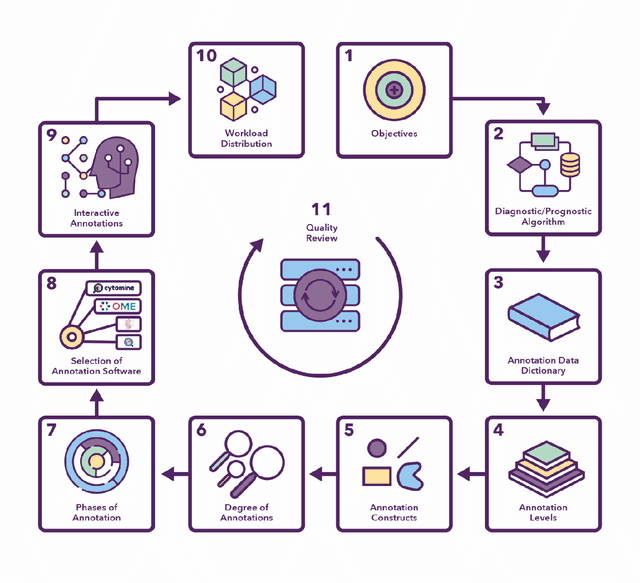
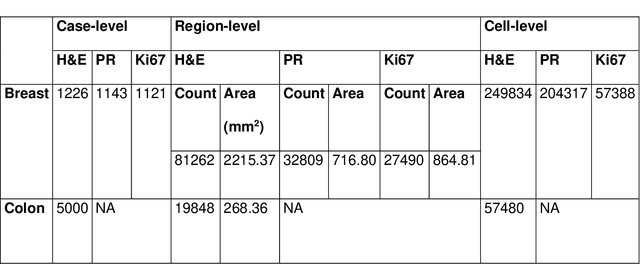
Abstract:Recent advances in whole slide imaging (WSI) technology have led to the development of a myriad of computer vision and artificial intelligence (AI) based diagnostic, prognostic, and predictive algorithms. Computational Pathology (CPath) offers an integrated solution to utilize information embedded in pathology WSIs beyond what we obtain through visual assessment. For automated analysis of WSIs and validation of machine learning (ML) models, annotations at the slide, tissue and cellular levels are required. The annotation of important visual constructs in pathology images is an important component of CPath projects. Improper annotations can result in algorithms which are hard to interpret and can potentially produce inaccurate and inconsistent results. Despite the crucial role of annotations in CPath projects, there are no well-defined guidelines or best practices on how annotations should be carried out. In this paper, we address this shortcoming by presenting the experience and best practices acquired during the execution of a large-scale annotation exercise involving a multidisciplinary team of pathologists, ML experts and researchers as part of the Pathology image data Lake for Analytics, Knowledge and Education (PathLAKE) consortium. We present a real-world case study along with examples of different types of annotations, diagnostic algorithm, annotation data dictionary and annotation constructs. The analyses reported in this work highlight best practice recommendations that can be used as annotation guidelines over the lifecycle of a CPath project.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge