Mohsin Bilal
Foundation Models in Computational Pathology: A Review of Challenges, Opportunities, and Impact
Feb 12, 2025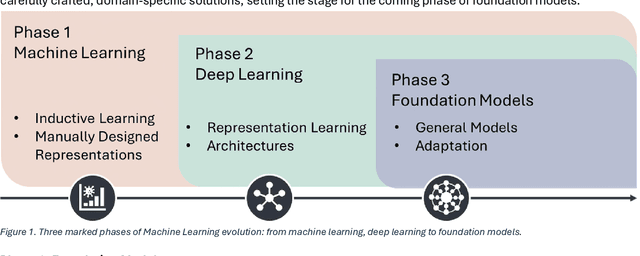
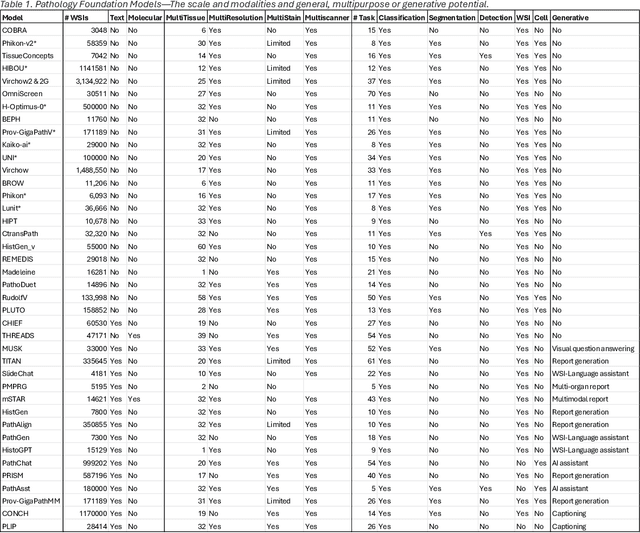
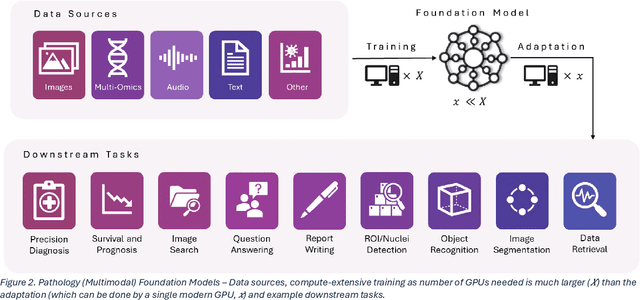
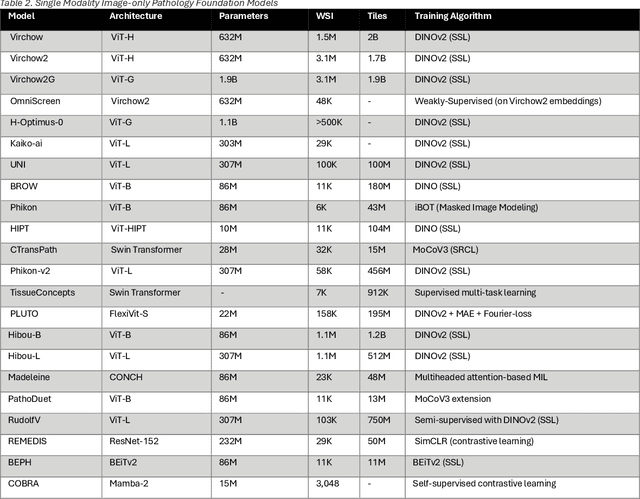
Abstract:From self-supervised, vision-only models to contrastive visual-language frameworks, computational pathology has rapidly evolved in recent years. Generative AI "co-pilots" now demonstrate the ability to mine subtle, sub-visual tissue cues across the cellular-to-pathology spectrum, generate comprehensive reports, and respond to complex user queries. The scale of data has surged dramatically, growing from tens to millions of multi-gigapixel tissue images, while the number of trainable parameters in these models has risen to several billion. The critical question remains: how will this new wave of generative and multi-purpose AI transform clinical diagnostics? In this article, we explore the true potential of these innovations and their integration into clinical practice. We review the rapid progress of foundation models in pathology, clarify their applications and significance. More precisely, we examine the very definition of foundational models, identifying what makes them foundational, general, or multipurpose, and assess their impact on computational pathology. Additionally, we address the unique challenges associated with their development and evaluation. These models have demonstrated exceptional predictive and generative capabilities, but establishing global benchmarks is crucial to enhancing evaluation standards and fostering their widespread clinical adoption. In computational pathology, the broader impact of frontier AI ultimately depends on widespread adoption and societal acceptance. While direct public exposure is not strictly necessary, it remains a powerful tool for dispelling misconceptions, building trust, and securing regulatory support.
Intelligent Known and Novel Aircraft Recognition -- A Shift from Classification to Similarity Learning for Combat Identification
Feb 26, 2024



Abstract:Precise aircraft recognition in low-resolution remote sensing imagery is a challenging yet crucial task in aviation, especially combat identification. This research addresses this problem with a novel, scalable, and AI-driven solution. The primary hurdle in combat identification in remote sensing imagery is the accurate recognition of Novel/Unknown types of aircraft in addition to Known types. Traditional methods, human expert-driven combat identification and image classification, fall short in identifying Novel classes. Our methodology employs similarity learning to discern features of a broad spectrum of military and civilian aircraft. It discerns both Known and Novel aircraft types, leveraging metric learning for the identification and supervised few-shot learning for aircraft type classification. To counter the challenge of limited low-resolution remote sensing data, we propose an end-to-end framework that adapts to the diverse and versatile process of military aircraft recognition by training a generalized embedder in fully supervised manner. Comparative analysis with earlier aircraft image classification methods shows that our approach is effective for aircraft image classification (F1-score Aircraft Type of 0.861) and pioneering for quantifying the identification of Novel types (F1-score Bipartitioning of 0.936). The proposed methodology effectively addresses inherent challenges in remote sensing data, thereby setting new standards in dataset quality. The research opens new avenues for domain experts and demonstrates unique capabilities in distinguishing various aircraft types, contributing to a more robust, domain-adapted potential for real-time aircraft recognition.
An Aggregation of Aggregation Methods in Computational Pathology
Nov 02, 2022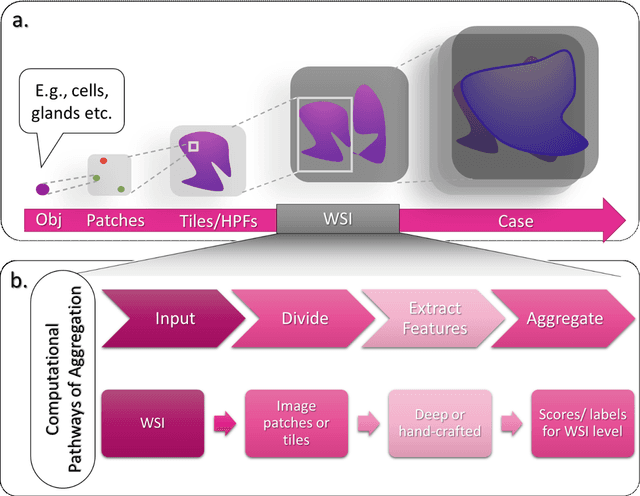
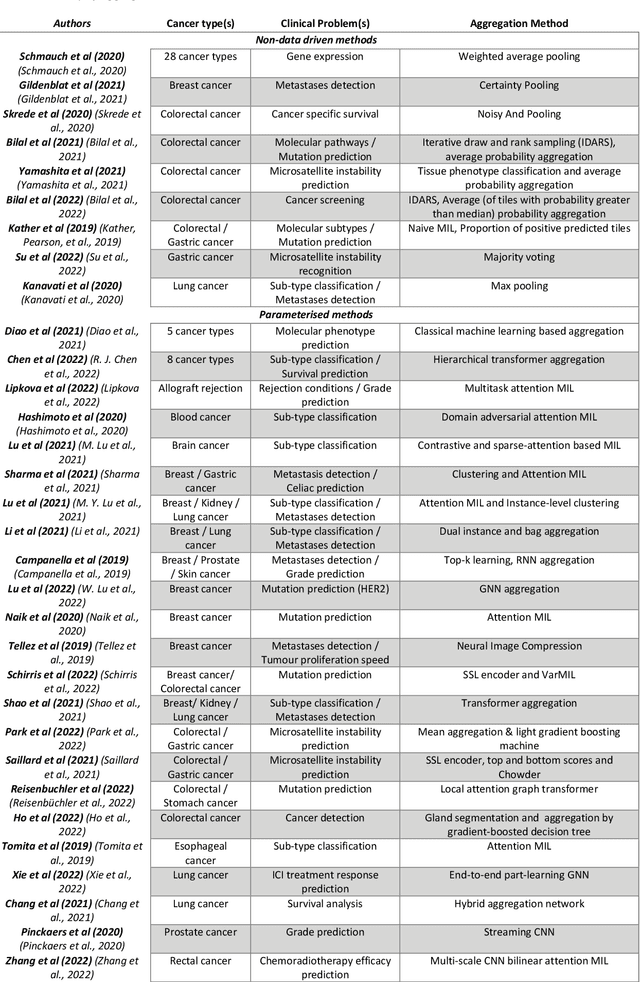
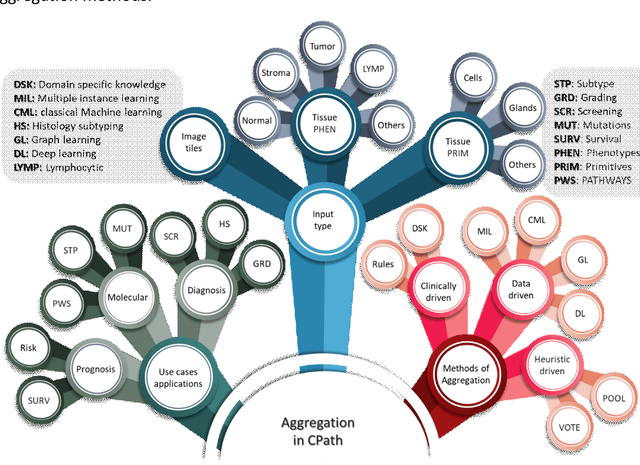
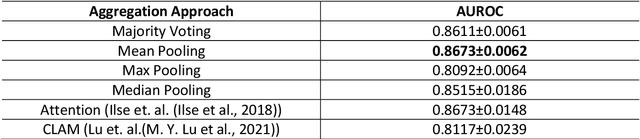
Abstract:Image analysis and machine learning algorithms operating on multi-gigapixel whole-slide images (WSIs) often process a large number of tiles (sub-images) and require aggregating predictions from the tiles in order to predict WSI-level labels. In this paper, we present a review of existing literature on various types of aggregation methods with a view to help guide future research in the area of computational pathology (CPath). We propose a general CPath workflow with three pathways that consider multiple levels and types of data and the nature of computation to analyse WSIs for predictive modelling. We categorize aggregation methods according to the context and representation of the data, features of computational modules and CPath use cases. We compare and contrast different methods based on the principle of multiple instance learning, perhaps the most commonly used aggregation method, covering a wide range of CPath literature. To provide a fair comparison, we consider a specific WSI-level prediction task and compare various aggregation methods for that task. Finally, we conclude with a list of objectives and desirable attributes of aggregation methods in general, pros and cons of the various approaches, some recommendations and possible future directions.
FedDropoutAvg: Generalizable federated learning for histopathology image classification
Nov 25, 2021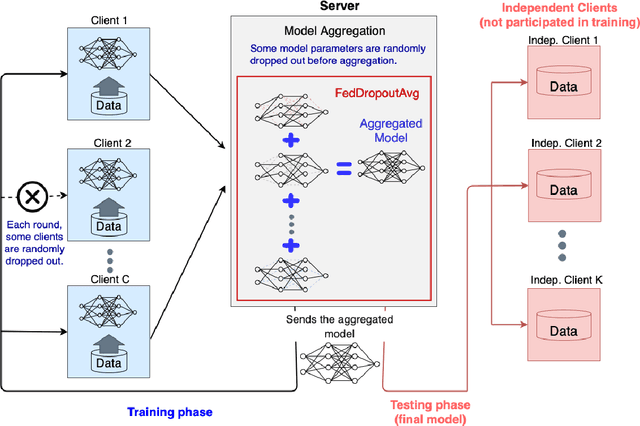
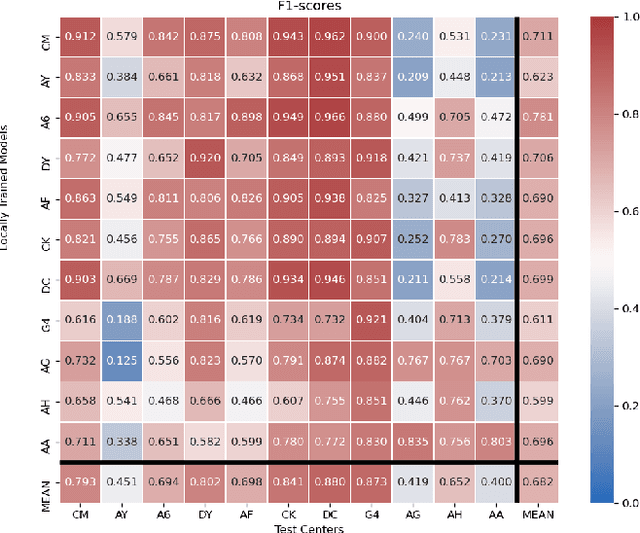
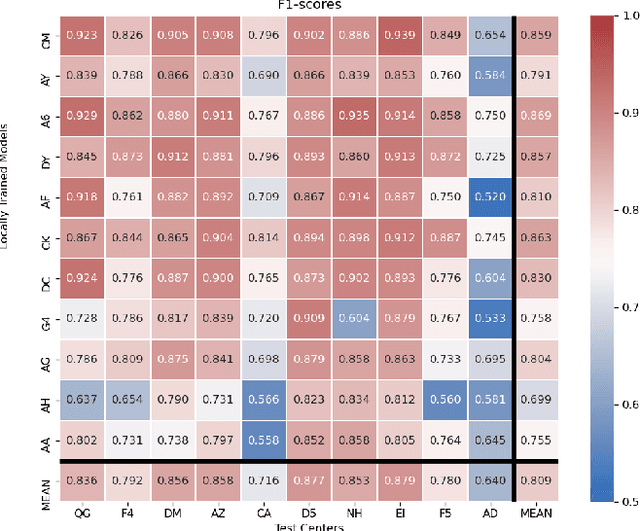
Abstract:Federated learning (FL) enables collaborative learning of a deep learning model without sharing the data of participating sites. FL in medical image analysis tasks is relatively new and open for enhancements. In this study, we propose FedDropoutAvg, a new federated learning approach for training a generalizable model. The proposed method takes advantage of randomness, both in client selection and also in federated averaging process. We compare FedDropoutAvg to several algorithms in an FL scenario for real-world multi-site histopathology image classification task. We show that with FedDropoutAvg, the final model can achieve performance better than other FL approaches and closer to a classical deep learning model that requires all data to be shared for centralized training. We test the trained models on a large dataset consisting of 1.2 million image tiles from 21 different centers. To evaluate the generalization ability of the proposed approach, we use held-out test sets from centers whose data was used in the FL and for unseen data from other independent centers whose data was not used in the federated training. We show that the proposed approach is more generalizable than other state-of-the-art federated training approaches. To the best of our knowledge, ours is the first study to use a randomized client and local model parameter selection procedure in a federated setting for a medical image analysis task.
Stain-Robust Mitotic Figure Detection for the Mitosis Domain Generalization Challenge
Sep 29, 2021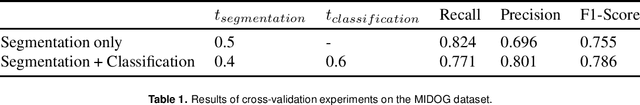
Abstract:The detection of mitotic figures from different scanners/sites remains an important topic of research, owing to its potential in assisting clinicians with tumour grading. The MItosis DOmain Generalization (MIDOG) challenge aims to test the robustness of detection models on unseen data from multiple scanners for this task. We present a short summary of the approach employed by the TIA Centre team to address this challenge. Our approach is based on a hybrid detection model, where mitotic candidates are segmented on stain normalised images, before being refined by a deep learning classifier. Cross-validation on the training images achieved the F1-score of 0.786 and 0.765 on the preliminary test set, demonstrating the generalizability of our model to unseen data from new scanners.
Semantic annotation for computational pathology: Multidisciplinary experience and best practice recommendations
Jun 25, 2021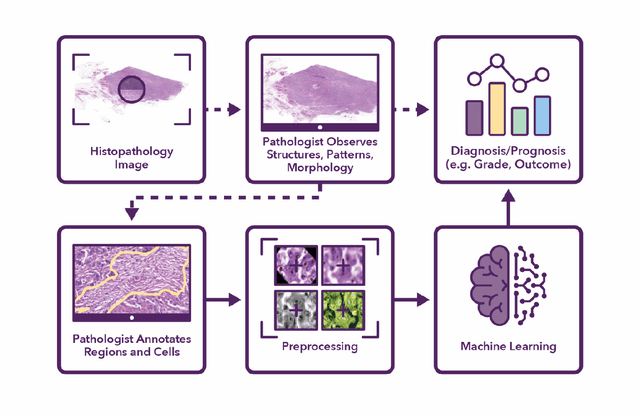
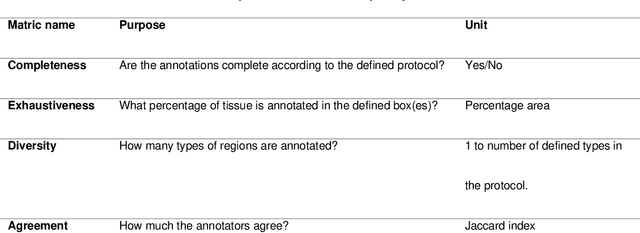
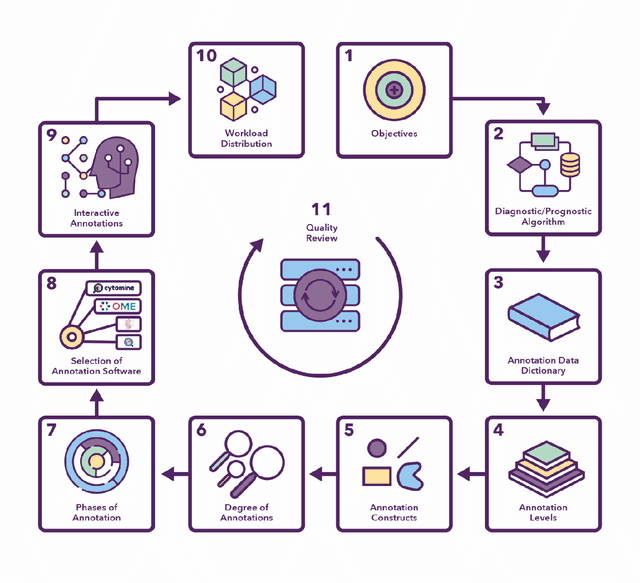
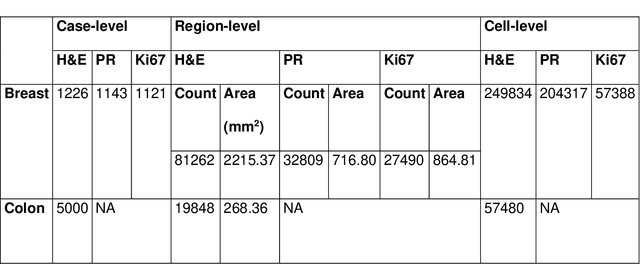
Abstract:Recent advances in whole slide imaging (WSI) technology have led to the development of a myriad of computer vision and artificial intelligence (AI) based diagnostic, prognostic, and predictive algorithms. Computational Pathology (CPath) offers an integrated solution to utilize information embedded in pathology WSIs beyond what we obtain through visual assessment. For automated analysis of WSIs and validation of machine learning (ML) models, annotations at the slide, tissue and cellular levels are required. The annotation of important visual constructs in pathology images is an important component of CPath projects. Improper annotations can result in algorithms which are hard to interpret and can potentially produce inaccurate and inconsistent results. Despite the crucial role of annotations in CPath projects, there are no well-defined guidelines or best practices on how annotations should be carried out. In this paper, we address this shortcoming by presenting the experience and best practices acquired during the execution of a large-scale annotation exercise involving a multidisciplinary team of pathologists, ML experts and researchers as part of the Pathology image data Lake for Analytics, Knowledge and Education (PathLAKE) consortium. We present a real-world case study along with examples of different types of annotations, diagnostic algorithm, annotation data dictionary and annotation constructs. The analyses reported in this work highlight best practice recommendations that can be used as annotation guidelines over the lifecycle of a CPath project.
Deep Multi-Resolution Dictionary Learning for Histopathology Image Analysis
Apr 01, 2021



Abstract:The problem of recognizing various types of tissues present in multi-gigapixel histology images is an important fundamental pre-requisite for downstream analysis of the tumor microenvironment in a bottom-up analysis paradigm for computational pathology. In this paper, we propose a deep dictionary learning approach to solve the problem of tissue phenotyping in histology images. We propose deep Multi-Resolution Dictionary Learning (deepMRDL) in order to benefit from deep texture descriptors at multiple different spatial resolutions. We show the efficacy of the proposed approach through extensive experiments on four benchmark histology image datasets from different organs (colorectal cancer, breast cancer and breast lymphnodes) and tasks (namely, cancer grading, tissue phenotyping, tumor detection and tissue type classification). We also show that the proposed framework can employ most off-the-shelf CNNs models to generate effective deep texture descriptors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge