Nima Hatami
A Robust Ensemble Algorithm for Ischemic Stroke Lesion Segmentation: Generalizability and Clinical Utility Beyond the ISLES Challenge
Apr 03, 2024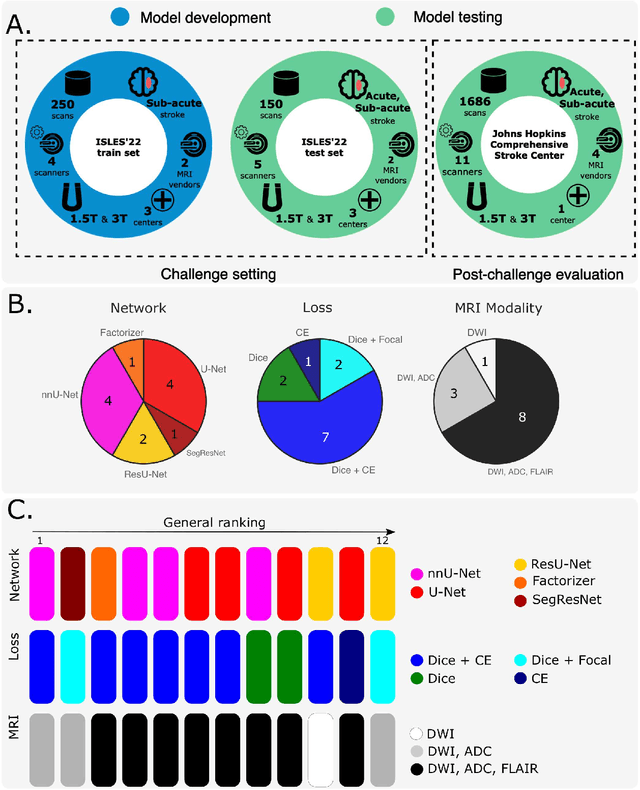
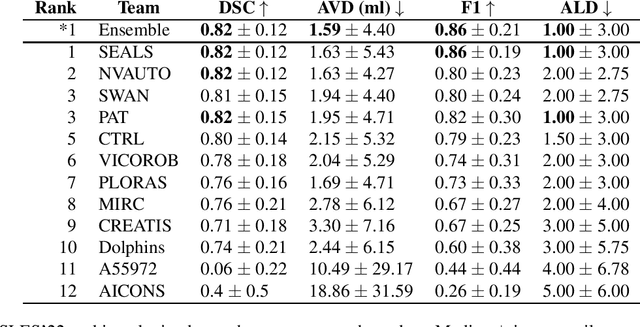
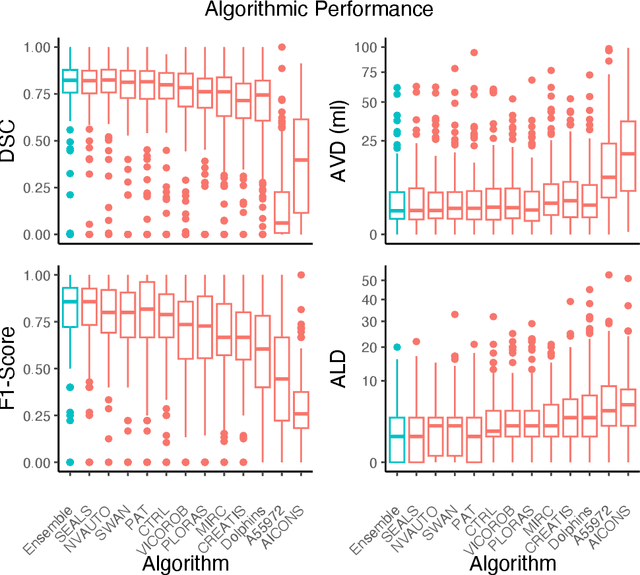
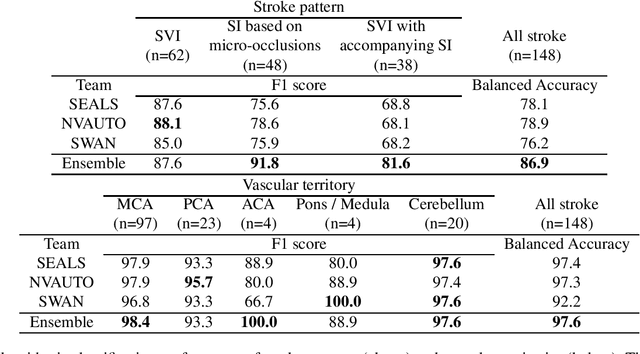
Abstract:Diffusion-weighted MRI (DWI) is essential for stroke diagnosis, treatment decisions, and prognosis. However, image and disease variability hinder the development of generalizable AI algorithms with clinical value. We address this gap by presenting a novel ensemble algorithm derived from the 2022 Ischemic Stroke Lesion Segmentation (ISLES) challenge. ISLES'22 provided 400 patient scans with ischemic stroke from various medical centers, facilitating the development of a wide range of cutting-edge segmentation algorithms by the research community. Through collaboration with leading teams, we combined top-performing algorithms into an ensemble model that overcomes the limitations of individual solutions. Our ensemble model achieved superior ischemic lesion detection and segmentation accuracy on our internal test set compared to individual algorithms. This accuracy generalized well across diverse image and disease variables. Furthermore, the model excelled in extracting clinical biomarkers. Notably, in a Turing-like test, neuroradiologists consistently preferred the algorithm's segmentations over manual expert efforts, highlighting increased comprehensiveness and precision. Validation using a real-world external dataset (N=1686) confirmed the model's generalizability. The algorithm's outputs also demonstrated strong correlations with clinical scores (admission NIHSS and 90-day mRS) on par with or exceeding expert-derived results, underlining its clinical relevance. This study offers two key findings. First, we present an ensemble algorithm (https://github.com/Tabrisrei/ISLES22_Ensemble) that detects and segments ischemic stroke lesions on DWI across diverse scenarios on par with expert (neuro)radiologists. Second, we show the potential for biomedical challenge outputs to extend beyond the challenge's initial objectives, demonstrating their real-world clinical applicability.
A Novel Autoencoders-LSTM Model for Stroke Outcome Prediction using Multimodal MRI Data
Mar 16, 2023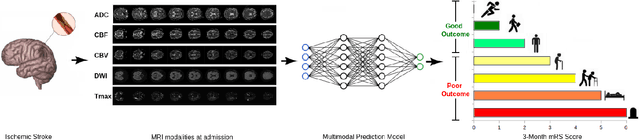
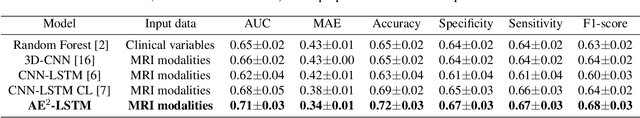
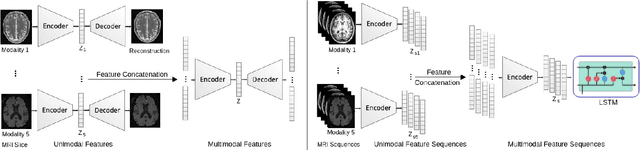
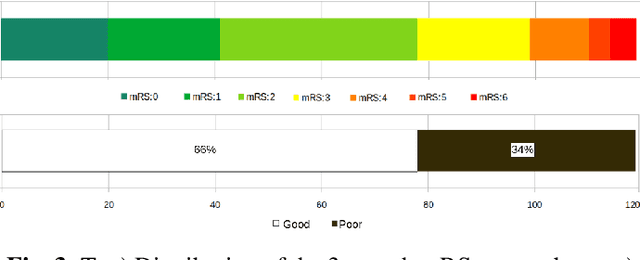
Abstract:Patient outcome prediction is critical in management of ischemic stroke. In this paper, a novel machine learning model is proposed for stroke outcome prediction using multimodal Magnetic Resonance Imaging (MRI). The proposed model consists of two serial levels of Autoencoders (AEs), where different AEs at level 1 are used for learning unimodal features from different MRI modalities and a AE at level 2 is used to combine the unimodal features into compressed multimodal features. The sequences of multimodal features of a given patient are then used by an LSTM network for predicting outcome score. The proposed AE2-LSTM model is proved to be an effective approach for better addressing the multimodality and volumetric nature of MRI data. Experimental results show that the proposed AE2-LSTM outperforms the existing state-of-the art models by achieving highest AUC=0.71 and lowest MAE=0.34.
DeepFEL: Deep Fastfood Ensemble Learning for Histopathology Image Analysis
Jan 23, 2023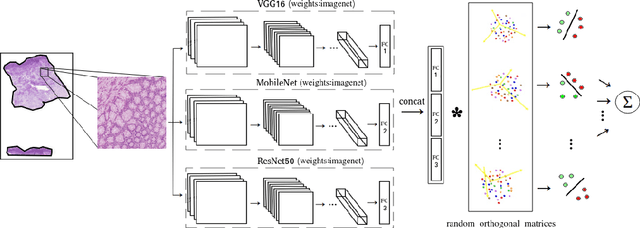
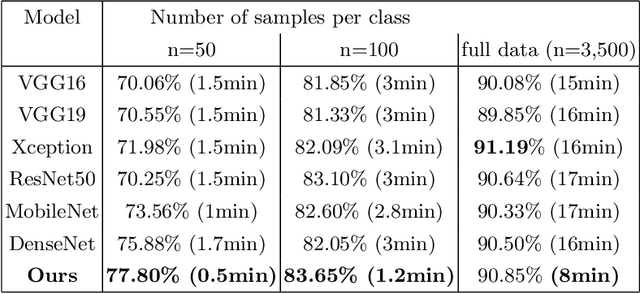
Abstract:Computational pathology tasks have some unique characterises such as multi-gigapixel images, tedious and frequently uncertain annotations, and unavailability of large number of cases [13]. To address some of these issues, we present Deep Fastfood Ensembles - a simple, fast and yet effective method for combining deep features pooled from popular CNN models pre-trained on totally different source domains (e.g., natural image objects) and projected onto diverse dimensions using random projections, the so-called Fastfood [11]. The final ensemble output is obtained by a consensus of simple individual classifiers, each of which is trained on a different collection of random basis vectors. This offers extremely fast and yet effective solution, especially when training times and domain labels are of the essence. We demonstrate the effectiveness of the proposed deep fastfood ensemble learning as compared to the state-of-the-art methods for three different tasks in histopathology image analysis.
CNN-LSTM Based Multimodal MRI and Clinical Data Fusion for Predicting Functional Outcome in Stroke Patients
May 11, 2022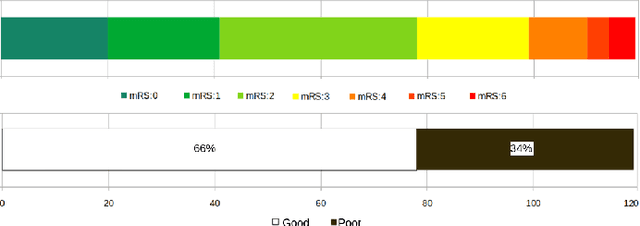
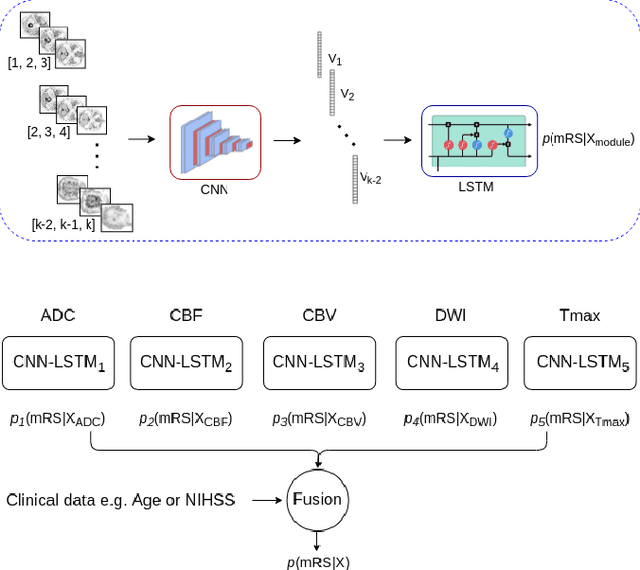
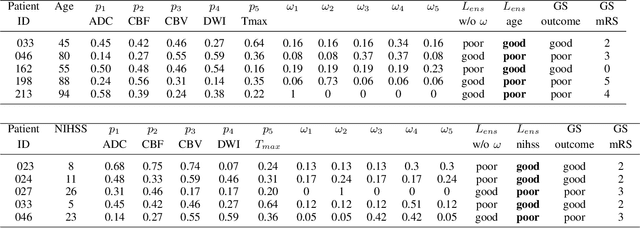
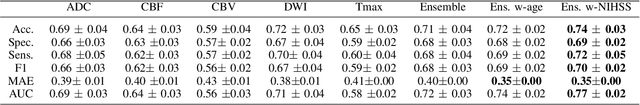
Abstract:Clinical outcome prediction plays an important role in stroke patient management. From a machine learning point-of-view, one of the main challenges is dealing with heterogeneous data at patient admission, i.e. the image data which are multidimensional and the clinical data which are scalars. In this paper, a multimodal convolutional neural network - long short-term memory (CNN-LSTM) based ensemble model is proposed. For each MR image module, a dedicated network provides preliminary prediction of the clinical outcome using the modified Rankin scale (mRS). The final mRS score is obtained by merging the preliminary probabilities of each module dedicated to a specific type of MR image weighted by the clinical metadata, here age or the National Institutes of Health Stroke Scale (NIHSS). The experimental results demonstrate that the proposed model surpasses the baselines and offers an original way to automatically encode the spatio-temporal context of MR images in a deep learning architecture. The highest AUC (0.77) was achieved for the proposed model with NIHSS.
Deep Multi-Resolution Dictionary Learning for Histopathology Image Analysis
Apr 01, 2021



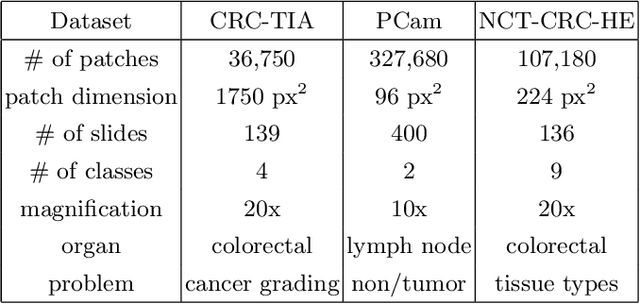
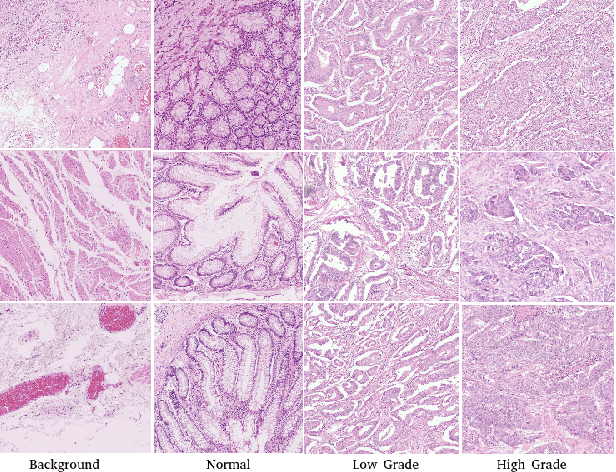
Abstract:The problem of recognizing various types of tissues present in multi-gigapixel histology images is an important fundamental pre-requisite for downstream analysis of the tumor microenvironment in a bottom-up analysis paradigm for computational pathology. In this paper, we propose a deep dictionary learning approach to solve the problem of tissue phenotyping in histology images. We propose deep Multi-Resolution Dictionary Learning (deepMRDL) in order to benefit from deep texture descriptors at multiple different spatial resolutions. We show the efficacy of the proposed approach through extensive experiments on four benchmark histology image datasets from different organs (colorectal cancer, breast cancer and breast lymphnodes) and tasks (namely, cancer grading, tissue phenotyping, tumor detection and tissue type classification). We also show that the proposed framework can employ most off-the-shelf CNNs models to generate effective deep texture descriptors.
Magnetic Resonance Spectroscopy Quantification using Deep Learning
Jun 19, 2018



Abstract:Magnetic resonance spectroscopy (MRS) is an important technique in biomedical research and it has the unique capability to give a non-invasive access to the biochemical content (metabolites) of scanned organs. In the literature, the quantification (the extraction of the potential biomarkers from the MRS signals) involves the resolution of an inverse problem based on a parametric model of the metabolite signal. However, poor signal-to-noise ratio (SNR), presence of the macromolecule signal or high correlation between metabolite spectral patterns can cause high uncertainties for most of the metabolites, which is one of the main reasons that prevents use of MRS in clinical routine. In this paper, quantification of metabolites in MR Spectroscopic imaging using deep learning is proposed. A regression framework based on the Convolutional Neural Networks (CNN) is introduced for an accurate estimation of spectral parameters. The proposed model learns the spectral features from a large-scale simulated data set with different variations of human brain spectra and SNRs. Experimental results demonstrate the accuracy of the proposed method, compared to state of the art standard quantification method (QUEST), on concentration of 20 metabolites and the macromolecule.
Bag of Recurrence Patterns Representation for Time-Series Classification
Mar 29, 2018



Abstract:Time-Series Classification (TSC) has attracted a lot of attention in pattern recognition, because wide range of applications from different domains such as finance and health informatics deal with time-series signals. Bag of Features (BoF) model has achieved a great success in TSC task by summarizing signals according to the frequencies of "feature words" of a data-learned dictionary. This paper proposes embedding the Recurrence Plots (RP), a visualization technique for analysis of dynamic systems, in the BoF model for TSC. While the traditional BoF approach extracts features from 1D signal segments, this paper uses the RP to transform time-series into 2D texture images and then applies the BoF on them. Image representation of time-series enables us to explore different visual descriptors that are not available for 1D signals and to treats TSC task as a texture recognition problem. Experimental results on the UCI time-series classification archive demonstrates a significant accuracy boost by the proposed Bag of Recurrence patterns (BoR), compared not only to the existing BoF models, but also to the state-of-the art algorithms.
Classification of Time-Series Images Using Deep Convolutional Neural Networks
Oct 07, 2017Abstract:Convolutional Neural Networks (CNN) has achieved a great success in image recognition task by automatically learning a hierarchical feature representation from raw data. While the majority of Time-Series Classification (TSC) literature is focused on 1D signals, this paper uses Recurrence Plots (RP) to transform time-series into 2D texture images and then take advantage of the deep CNN classifier. Image representation of time-series introduces different feature types that are not available for 1D signals, and therefore TSC can be treated as texture image recognition task. CNN model also allows learning different levels of representations together with a classifier, jointly and automatically. Therefore, using RP and CNN in a unified framework is expected to boost the recognition rate of TSC. Experimental results on the UCR time-series classification archive demonstrate competitive accuracy of the proposed approach, compared not only to the existing deep architectures, but also to the state-of-the art TSC algorithms.
Automatic Identification of Retinal Arteries and Veins in Fundus Images using Local Binary Patterns
May 03, 2016



Abstract:Artery and vein (AV) classification of retinal images is a key to necessary tasks, such as automated measurement of arteriolar-to-venular diameter ratio (AVR). This paper comprehensively reviews the state-of-the art in AV classification methods. To improve on previous methods, a new Local Bi- nary Pattern-based method (LBP) is proposed. Beside its simplicity, LBP is robust against low contrast and low quality fundus images; and it helps the process by including additional AV texture and shape information. Experimental results compare the performance of the new method with the state-of-the art; and also methods with different feature extraction and classification schemas.
ECOC-Based Training of Neural Networks for Face Recognition
Dec 14, 2013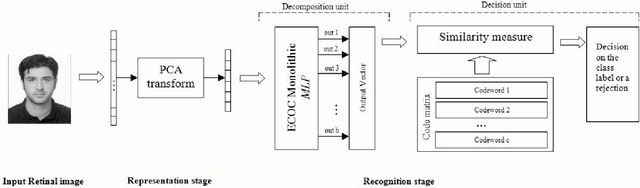

Abstract:Error Correcting Output Codes, ECOC, is an output representation method capable of discovering some of the errors produced in classification tasks. This paper describes the application of ECOC to the training of feed forward neural networks, FFNN, for improving the overall accuracy of classification systems. Indeed, to improve the generalization of FFNN classifiers, this paper proposes an ECOC-Based training method for Neural Networks that use ECOC as the output representation, and adopts the traditional Back-Propagation algorithm, BP, to adjust weights of the network. Experimental results for face recognition problem on Yale database demonstrate the effectiveness of our method. With a rejection scheme defined by a simple robustness rate, high reliability is achieved in this application.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge