David Rousseau
Unlocking Zero-Shot Plant Segmentation with Pl@ntNet Intelligence
Oct 14, 2025



Abstract:We present a zero-shot segmentation approach for agricultural imagery that leverages Plantnet, a large-scale plant classification model, in conjunction with its DinoV2 backbone and the Segment Anything Model (SAM). Rather than collecting and annotating new datasets, our method exploits Plantnet's specialized plant representations to identify plant regions and produce coarse segmentation masks. These masks are then refined by SAM to yield detailed segmentations. We evaluate on four publicly available datasets of various complexity in terms of contrast including some where the limited size of the training data and complex field conditions often hinder purely supervised methods. Our results show consistent performance gains when using Plantnet-fine-tuned DinoV2 over the base DinoV2 model, as measured by the Jaccard Index (IoU). These findings highlight the potential of combining foundation models with specialized plant-centric models to alleviate the annotation bottleneck and enable effective segmentation in diverse agricultural scenarios.
FAIR Universe HiggsML Uncertainty Challenge Competition
Oct 03, 2024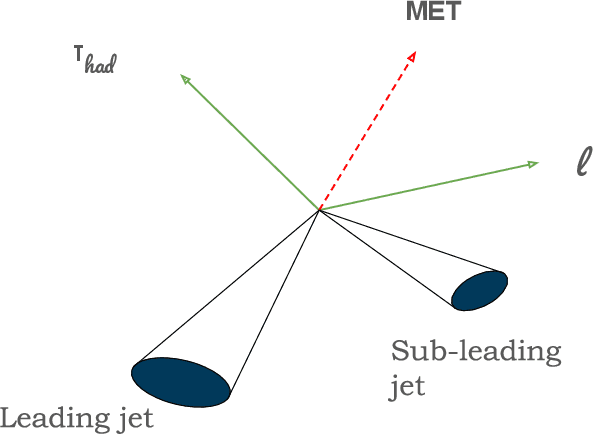
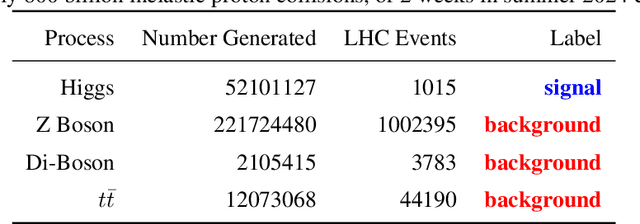
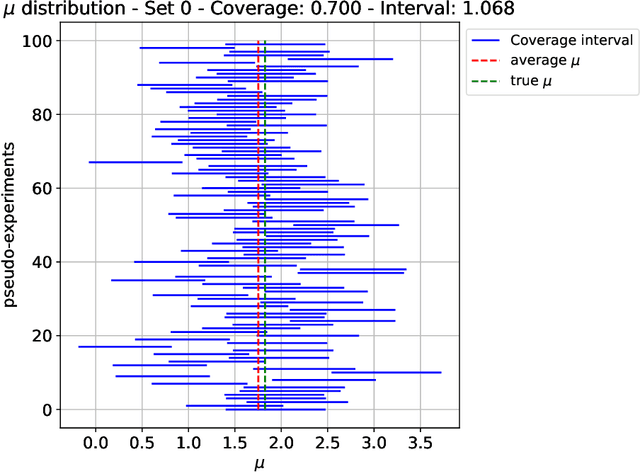
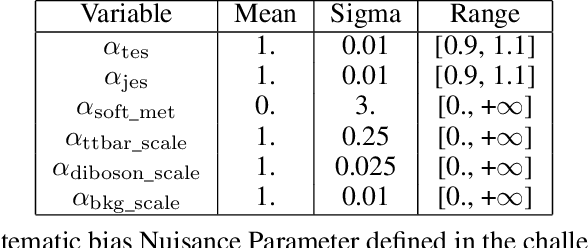
Abstract:The FAIR Universe -- HiggsML Uncertainty Challenge focuses on measuring the physics properties of elementary particles with imperfect simulators due to differences in modelling systematic errors. Additionally, the challenge is leveraging a large-compute-scale AI platform for sharing datasets, training models, and hosting machine learning competitions. Our challenge brings together the physics and machine learning communities to advance our understanding and methodologies in handling systematic (epistemic) uncertainties within AI techniques.
PLANesT-3D: A new annotated dataset for segmentation of 3D plant point clouds
Jul 30, 2024



Abstract:Creation of new annotated public datasets is crucial in helping advances in 3D computer vision and machine learning meet their full potential for automatic interpretation of 3D plant models. In this paper, we introduce PLANesT-3D; a new annotated dataset of 3D color point clouds of plants. PLANesT-3D is composed of 34 point cloud models representing 34 real plants from three different plant species: \textit{Capsicum annuum}, \textit{Rosa kordana}, and \textit{Ribes rubrum}. Both semantic labels in terms of "leaf" and "stem", and organ instance labels were manually annotated for the full point clouds. As an additional contribution, SP-LSCnet, a novel semantic segmentation method that is a combination of unsupervised superpoint extraction and a 3D point-based deep learning approach is introduced and evaluated on the new dataset. Two existing deep neural network architectures, PointNet++ and RoseSegNet were also tested on the point clouds of PLANesT-3D for semantic segmentation.
On-the-fly spectral unmixing based on Kalman filtering
Jul 22, 2024
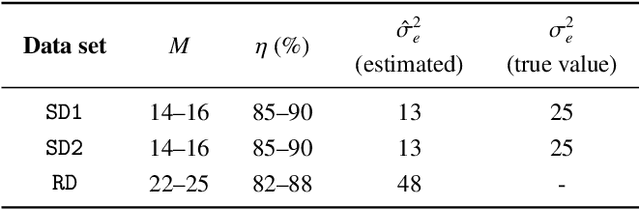
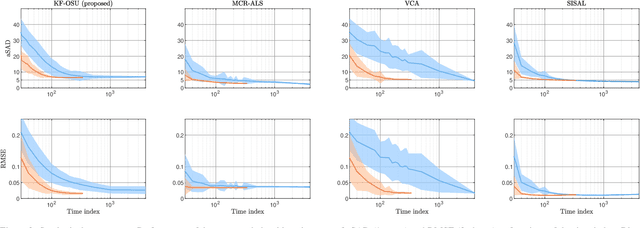
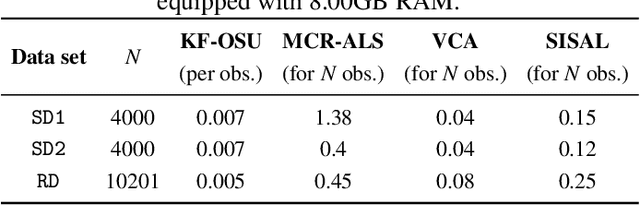
Abstract:This work introduces an on-the-fly (i.e., online) linear unmixing method which is able to sequentially analyze spectral data acquired on a spectrum-by-spectrum basis. After deriving a sequential counterpart of the conventional linear mixing model, the proposed approach recasts the linear unmixing problem into a linear state-space estimation framework. Under Gaussian noise and state models, the estimation of the pure spectra can be efficiently conducted by resorting to Kalman filtering. Interestingly, it is shown that this Kalman filter can operate in a lower-dimensional subspace while ensuring the nonnegativity constraint inherent to pure spectra. This dimensionality reduction allows significantly lightening the computational burden, while leveraging recent advances related to the representation of essential spectral information. The proposed method is evaluated through extensive numerical experiments conducted on synthetic and real Raman data sets. The results show that this Kalman filter-based method offers a convenient trade-off between unmixing accuracy and computational efficiency, which is crucial for operating in an on-the-fly setting. To the best of the authors' knowledge, this is the first operational method which is able to solve the spectral unmixing problem efficiently in a dynamic fashion. It also constitutes a valuable building block for benefiting from acquisition and processing frameworks recently proposed in the microscopy literature, which are motivated by practical issues such as reducing acquisition time and avoiding potential damages being inflicted to photosensitive samples.
AI Competitions and Benchmarks: towards impactful challenges with post-challenge papers, benchmarks and other dissemination actions
Dec 14, 2023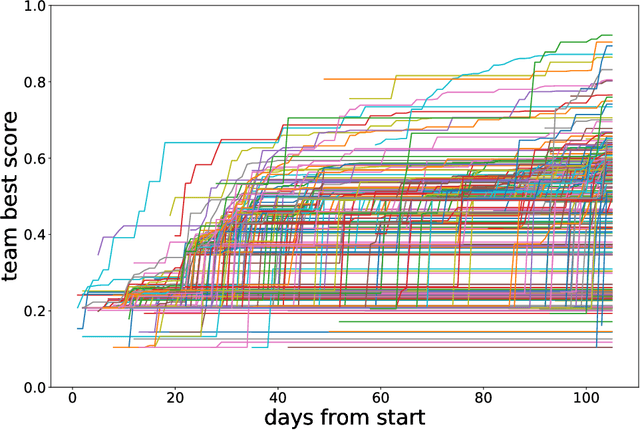
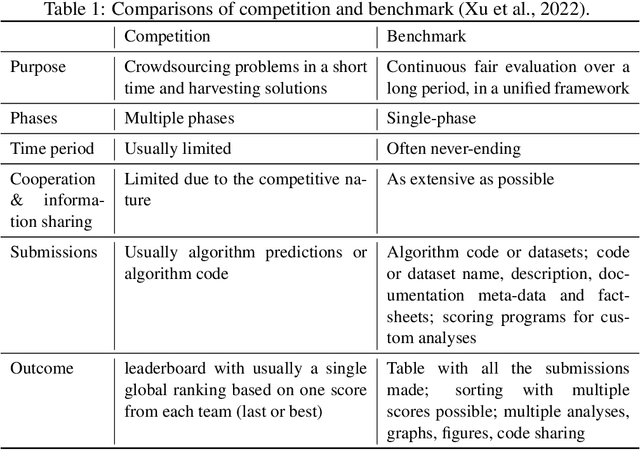
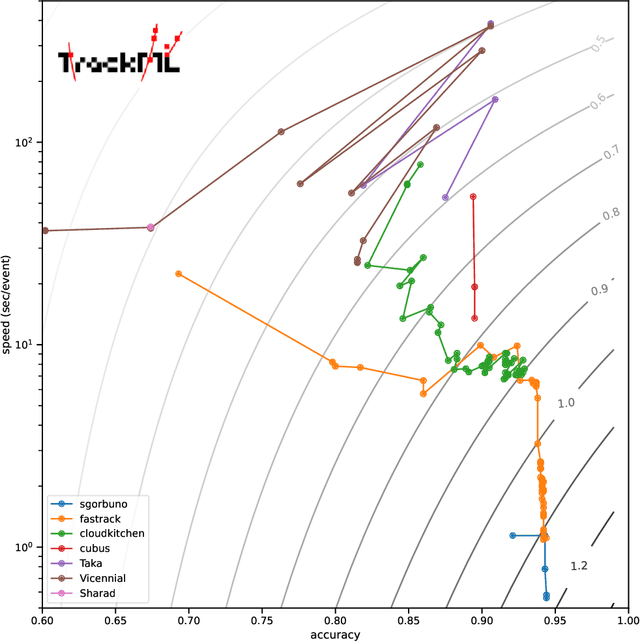
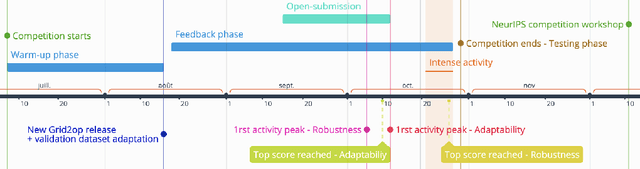
Abstract:Organising an AI challenge does not end with the final event. The long-lasting impact also needs to be organised. This chapter covers the various activities after the challenge is formally finished. The target audience of different post-challenge activities is identified. The various outputs of the challenge are listed with the means to collect them. The main part of the chapter is a template for a typical post-challenge paper, including possible graphs as well as advice on how to turn the challenge into a long-lasting benchmark.
Toward more frugal models for functional cerebral networks automatic recognition with resting-state fMRI
Jul 04, 2023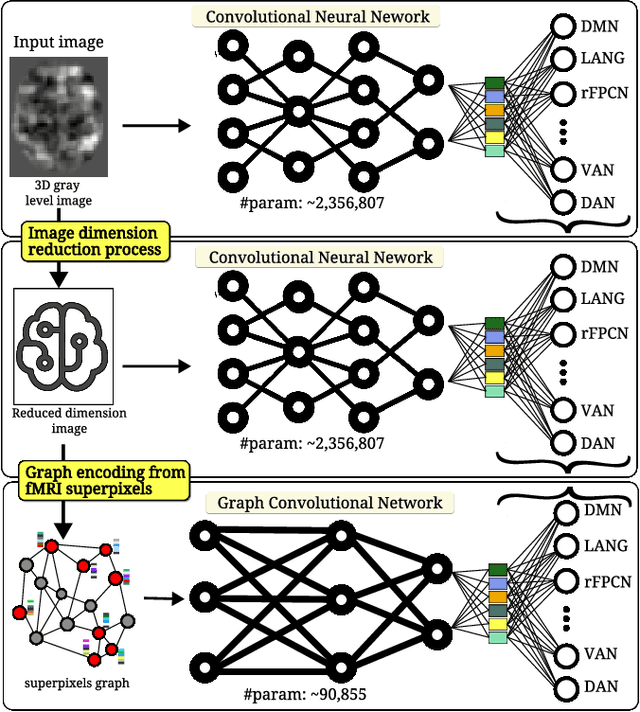
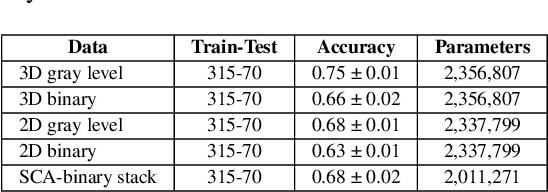
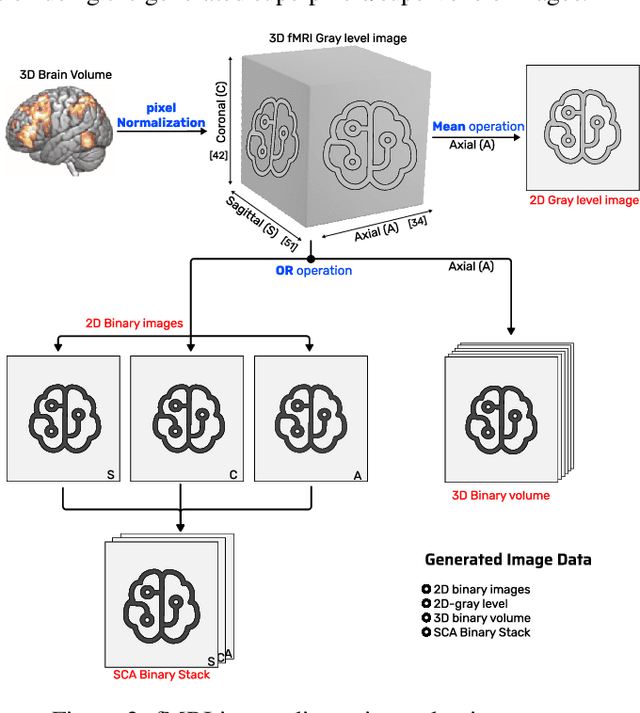
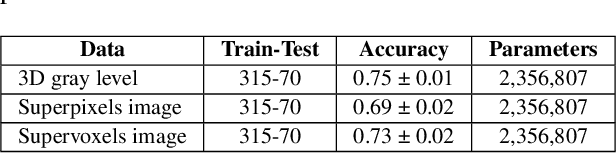
Abstract:We refer to a machine learning situation where models based on classical convolutional neural networks have shown good performance. We are investigating different encoding techniques in the form of supervoxels, then graphs to reduce the complexity of the model while tracking the loss of performance. This approach is illustrated on a recognition task of resting-state functional networks for patients with brain tumors. Graphs encoding supervoxels preserve activation characteristics of functional brain networks from images, optimize model parameters by 26 times while maintaining CNN model performance.
A Novel Autoencoders-LSTM Model for Stroke Outcome Prediction using Multimodal MRI Data
Mar 16, 2023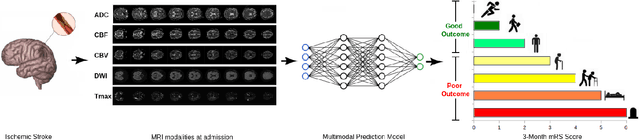
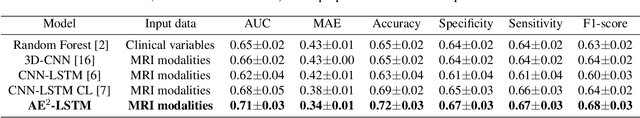
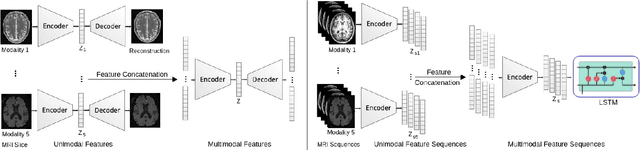
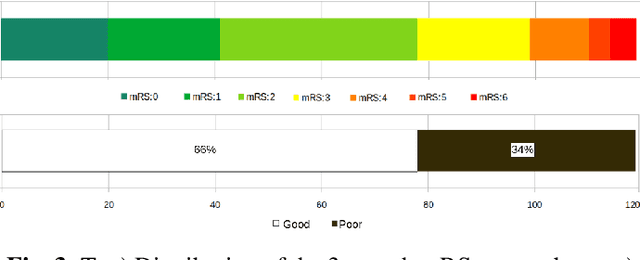
Abstract:Patient outcome prediction is critical in management of ischemic stroke. In this paper, a novel machine learning model is proposed for stroke outcome prediction using multimodal Magnetic Resonance Imaging (MRI). The proposed model consists of two serial levels of Autoencoders (AEs), where different AEs at level 1 are used for learning unimodal features from different MRI modalities and a AE at level 2 is used to combine the unimodal features into compressed multimodal features. The sequences of multimodal features of a given patient are then used by an LSTM network for predicting outcome score. The proposed AE2-LSTM model is proved to be an effective approach for better addressing the multimodality and volumetric nature of MRI data. Experimental results show that the proposed AE2-LSTM outperforms the existing state-of-the art models by achieving highest AUC=0.71 and lowest MAE=0.34.
CNN-LSTM Based Multimodal MRI and Clinical Data Fusion for Predicting Functional Outcome in Stroke Patients
May 11, 2022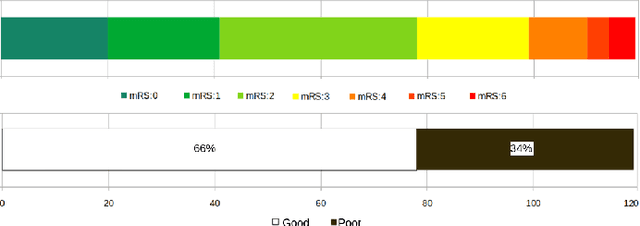
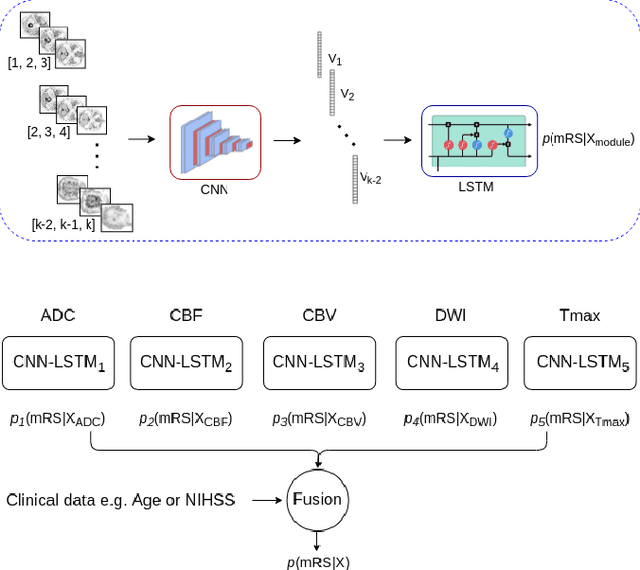
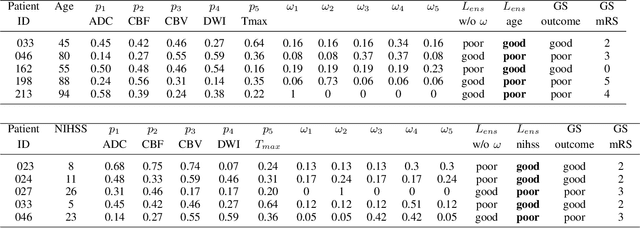
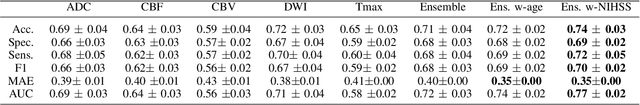
Abstract:Clinical outcome prediction plays an important role in stroke patient management. From a machine learning point-of-view, one of the main challenges is dealing with heterogeneous data at patient admission, i.e. the image data which are multidimensional and the clinical data which are scalars. In this paper, a multimodal convolutional neural network - long short-term memory (CNN-LSTM) based ensemble model is proposed. For each MR image module, a dedicated network provides preliminary prediction of the clinical outcome using the modified Rankin scale (mRS). The final mRS score is obtained by merging the preliminary probabilities of each module dedicated to a specific type of MR image weighted by the clinical metadata, here age or the National Institutes of Health Stroke Scale (NIHSS). The experimental results demonstrate that the proposed model surpasses the baselines and offers an original way to automatically encode the spatio-temporal context of MR images in a deep learning architecture. The highest AUC (0.77) was achieved for the proposed model with NIHSS.
The Tracking Machine Learning challenge : Throughput phase
May 14, 2021
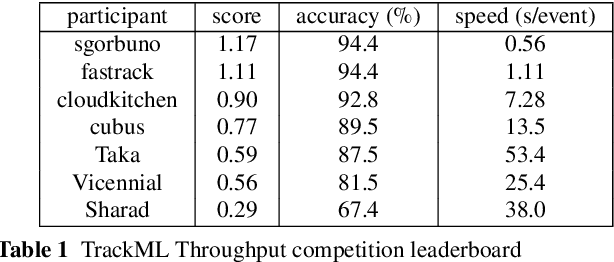
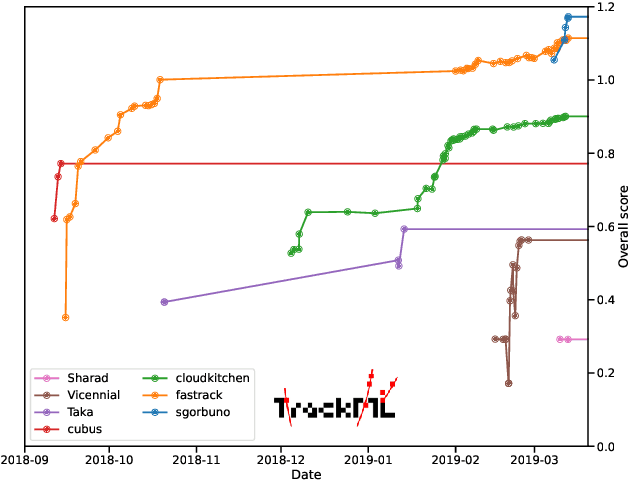
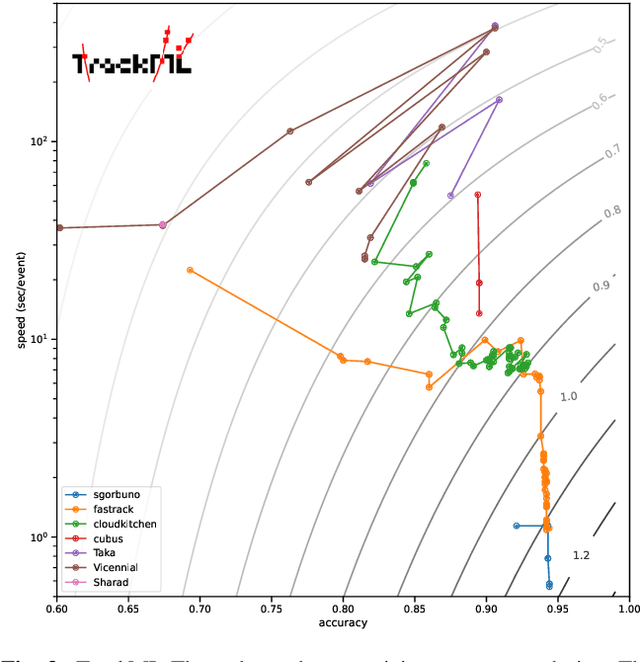
Abstract:This paper reports on the second "Throughput" phase of the Tracking Machine Learning (TrackML) challenge on the Codalab platform. As in the first "Accuracy" phase, the participants had to solve a difficult experimental problem linked to tracking accurately the trajectory of particles as e.g. created at the Large Hadron Collider (LHC): given O($10^5$) points, the participants had to connect them into O($10^4$) individual groups that represent the particle trajectories which are approximated helical. While in the first phase only the accuracy mattered, the goal of this second phase was a compromise between the accuracy and the speed of inference. Both were measured on the Codalab platform where the participants had to upload their software. The best three participants had solutions with good accuracy and speed an order of magnitude faster than the state of the art when the challenge was designed. Although the core algorithms were less diverse than in the first phase, a diversity of techniques have been used and are described in this paper. The performance of the algorithms are analysed in depth and lessons derived.
Assigning Apples to Individual Trees in Dense Orchards using 3D Color Point Clouds
Dec 26, 2020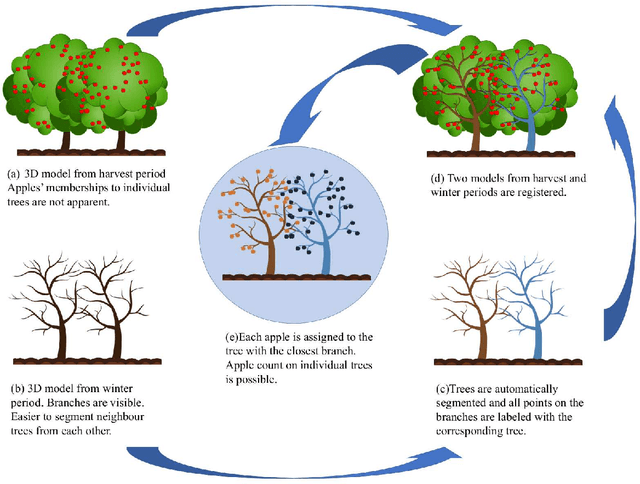
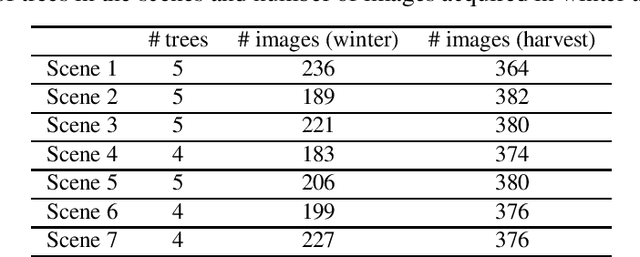
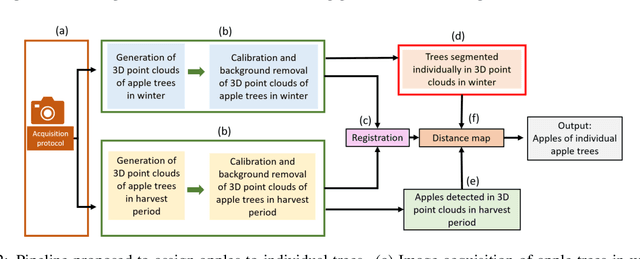
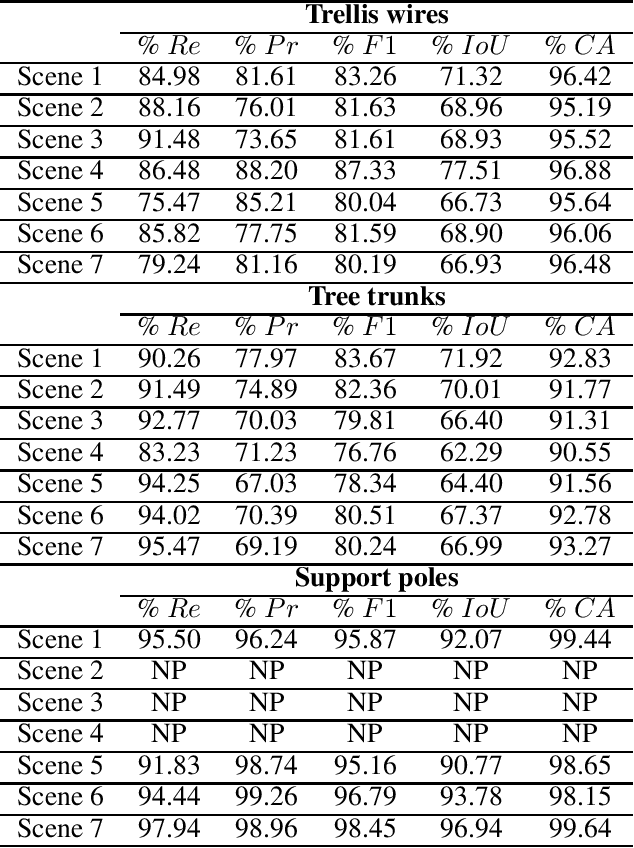
Abstract:We propose a 3D color point cloud processing pipeline to count apples on individual apple trees in trellis structured orchards. Fruit counting at the tree level requires separating trees, which is challenging in dense orchards. We employ point clouds acquired from the leaf-off orchard in winter period, where the branch structure is visible, to delineate tree crowns. We localize apples in point clouds acquired in harvest period. Alignment of the two point clouds enables mapping apple locations to the delineated winter cloud and assigning each apple to its bearing tree. Our apple assignment method achieves an accuracy rate higher than 95%. In addition to presenting a first proof of feasibility, we also provide suggestions for further improvement on our apple assignment pipeline.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge