R. M. Saad Bashir
Stain-Robust Mitotic Figure Detection for the Mitosis Domain Generalization Challenge
Sep 29, 2021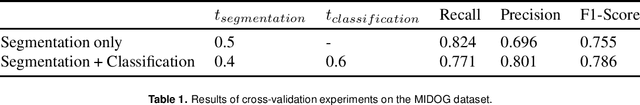
Abstract:The detection of mitotic figures from different scanners/sites remains an important topic of research, owing to its potential in assisting clinicians with tumour grading. The MItosis DOmain Generalization (MIDOG) challenge aims to test the robustness of detection models on unseen data from multiple scanners for this task. We present a short summary of the approach employed by the TIA Centre team to address this challenge. Our approach is based on a hybrid detection model, where mitotic candidates are segmented on stain normalised images, before being refined by a deep learning classifier. Cross-validation on the training images achieved the F1-score of 0.786 and 0.765 on the preliminary test set, demonstrating the generalizability of our model to unseen data from new scanners.
Simultaneous Nuclear Instance and Layer Segmentation in Oral Epithelial Dysplasia
Sep 01, 2021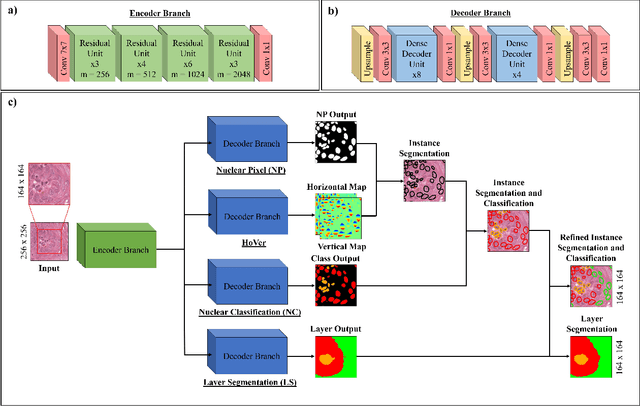
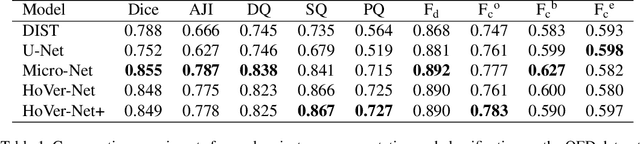
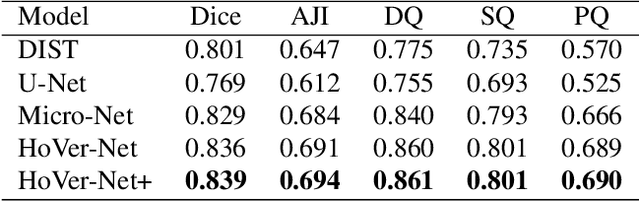
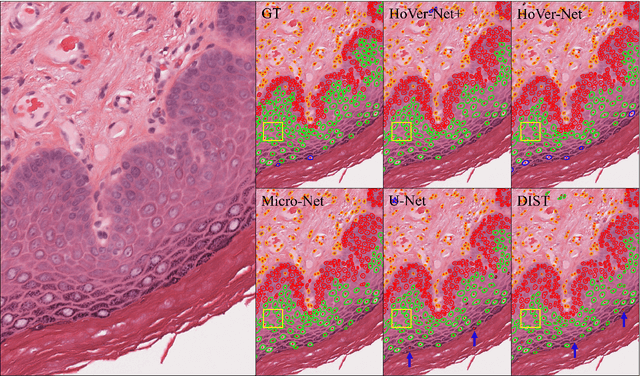
Abstract:Oral epithelial dysplasia (OED) is a pre-malignant histopathological diagnosis given to lesions of the oral cavity. Predicting OED grade or whether a case will transition to malignancy is critical for early detection and appropriate treatment. OED typically begins in the lower third of the epithelium before progressing upwards with grade severity, thus we have suggested that segmenting intra-epithelial layers, in addition to individual nuclei, may enable researchers to evaluate important layer-specific morphological features for grade/malignancy prediction. We present HoVer-Net+, a deep learning framework to simultaneously segment (and classify) nuclei and (intra-)epithelial layers in H&E stained slides from OED cases. The proposed architecture consists of an encoder branch and four decoder branches for simultaneous instance segmentation of nuclei and semantic segmentation of the epithelial layers. We show that the proposed model achieves the state-of-the-art (SOTA) performance in both tasks, with no additional costs when compared to previous SOTA methods for each task. To the best of our knowledge, ours is the first method for simultaneous nuclear instance segmentation and semantic tissue segmentation, with potential for use in computational pathology for other similar simultaneous tasks and for future studies into malignancy prediction.
HydraMix-Net: A Deep Multi-task Semi-supervised Learning Approach for Cell Detection and Classification
Aug 11, 2020Abstract:Semi-supervised techniques have removed the barriers of large scale labelled set by exploiting unlabelled data to improve the performance of a model. In this paper, we propose a semi-supervised deep multi-task classification and localization approach HydraMix-Net in the field of medical imagining where labelling is time consuming and costly. Firstly, the pseudo labels are generated using the model's prediction on the augmented set of unlabelled image with averaging. The high entropy predictions are further sharpened to reduced the entropy and are then mixed with the labelled set for training. The model is trained in multi-task learning manner with noise tolerant joint loss for classification localization and achieves better performance when given limited data in contrast to a simple deep model. On DLBCL data it achieves 80\% accuracy in contrast to simple CNN achieving 70\% accuracy when given only 100 labelled examples.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge