Hanya Mahmood
Transformer-based Model for Oral Epithelial Dysplasia Segmentation
Nov 09, 2023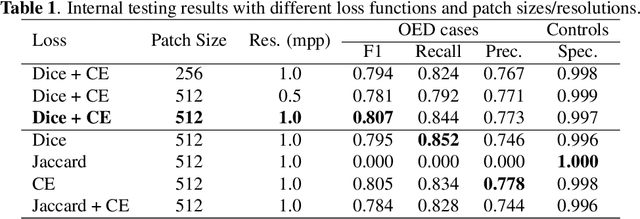
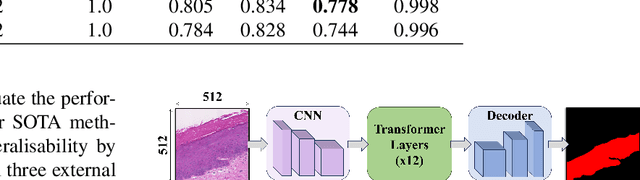
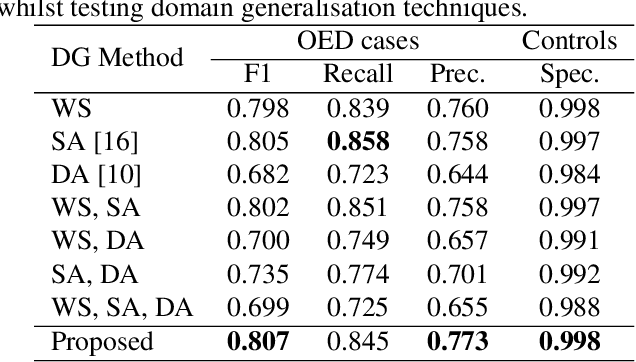
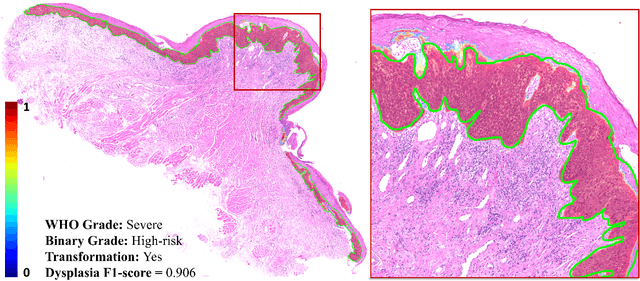
Abstract:Oral epithelial dysplasia (OED) is a premalignant histopathological diagnosis given to lesions of the oral cavity. OED grading is subject to large inter/intra-rater variability, resulting in the under/over-treatment of patients. We developed a new Transformer-based pipeline to improve detection and segmentation of OED in haematoxylin and eosin (H&E) stained whole slide images (WSIs). Our model was trained on OED cases (n = 260) and controls (n = 105) collected using three different scanners, and validated on test data from three external centres in the United Kingdom and Brazil (n = 78). Our internal experiments yield a mean F1-score of 0.81 for OED segmentation, which reduced slightly to 0.71 on external testing, showing good generalisability, and gaining state-of-the-art results. This is the first externally validated study to use Transformers for segmentation in precancerous histology images. Our publicly available model shows great promise to be the first step of a fully-integrated pipeline, allowing earlier and more efficient OED diagnosis, ultimately benefiting patient outcomes.
A Fully Automated and Explainable Algorithm for the Prediction of Malignant Transformation in Oral Epithelial Dysplasia
Jul 06, 2023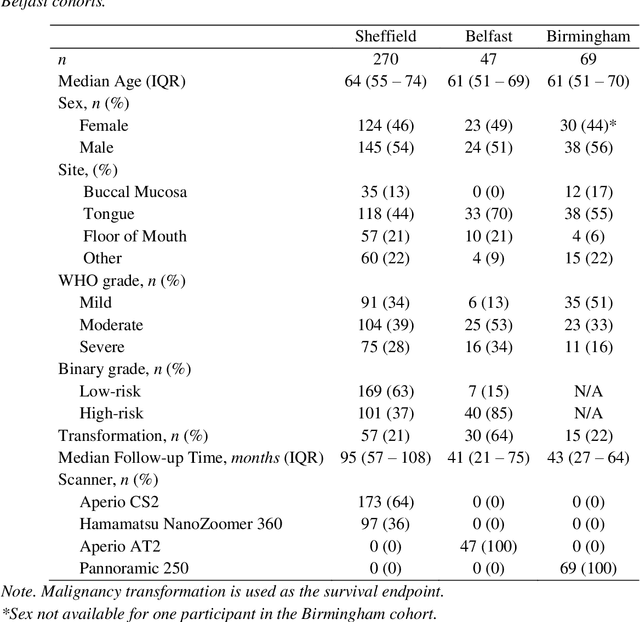
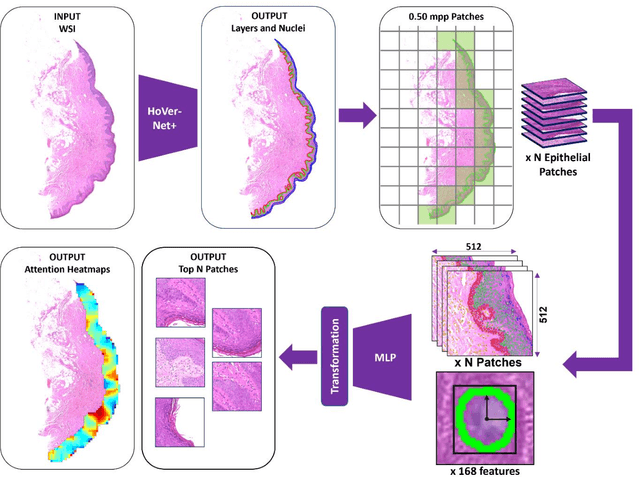
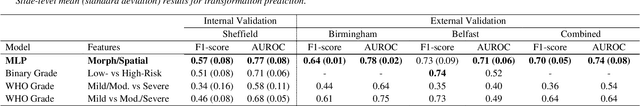
Abstract:Oral epithelial dysplasia (OED) is a premalignant histopathological diagnosis given to lesions of the oral cavity. Its grading suffers from significant inter-/intra- observer variability, and does not reliably predict malignancy progression, potentially leading to suboptimal treatment decisions. To address this, we developed a novel artificial intelligence algorithm that can assign an Oral Malignant Transformation (OMT) risk score, based on histological patterns in the in Haematoxylin and Eosin stained whole slide images, to quantify the risk of OED progression. The algorithm is based on the detection and segmentation of nuclei within (and around) the epithelium using an in-house segmentation model. We then employed a shallow neural network fed with interpretable morphological/spatial features, emulating histological markers. We conducted internal cross-validation on our development cohort (Sheffield; n = 193 cases) followed by independent validation on two external cohorts (Birmingham and Belfast; n = 92 cases). The proposed OMTscore yields an AUROC = 0.74 in predicting whether an OED progresses to malignancy or not. Survival analyses showed the prognostic value of our OMTscore for predicting malignancy transformation, when compared to the manually-assigned WHO and binary grades. Analysis of the correctly predicted cases elucidated the presence of peri-epithelial and epithelium-infiltrating lymphocytes in the most predictive patches of cases that transformed (p < 0.0001). This is the first study to propose a completely automated algorithm for predicting OED transformation based on interpretable nuclear features, whilst being validated on external datasets. The algorithm shows better-than-human-level performance for prediction of OED malignant transformation and offers a promising solution to the challenges of grading OED in routine clinical practice.
Simultaneous Nuclear Instance and Layer Segmentation in Oral Epithelial Dysplasia
Sep 01, 2021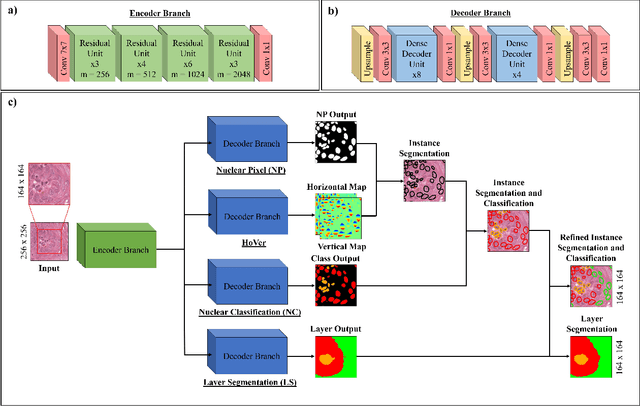
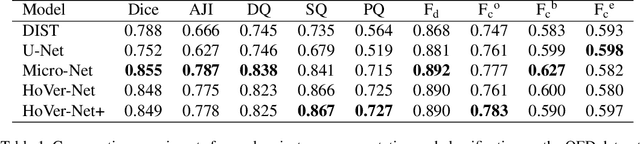
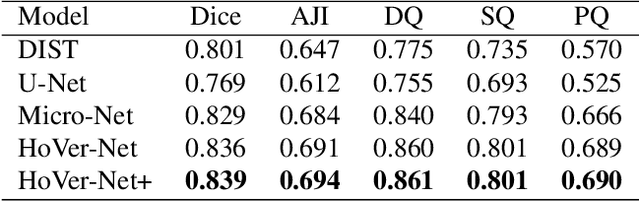
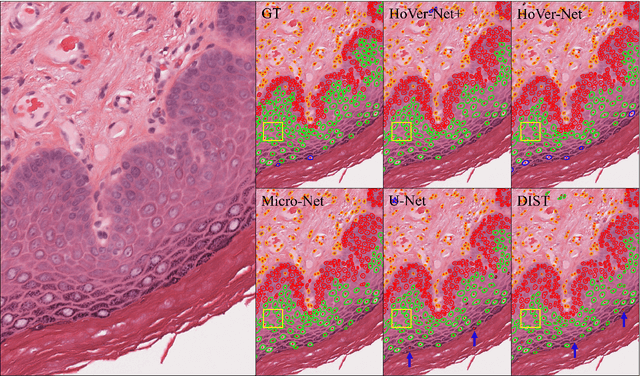
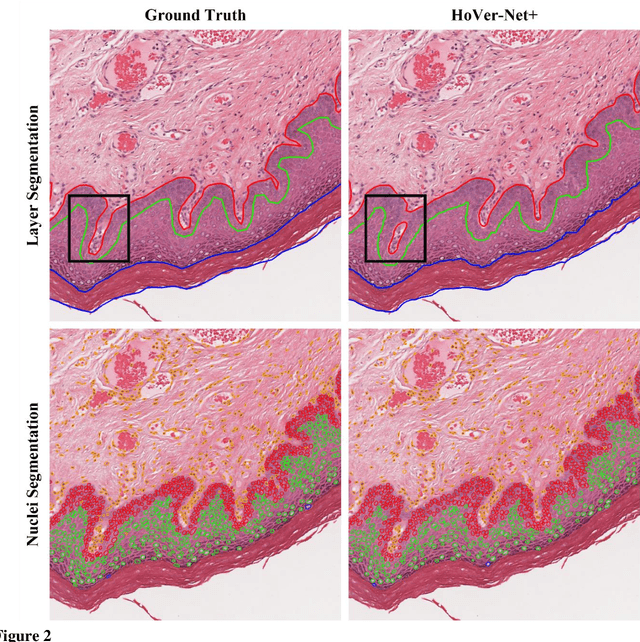
Abstract:Oral epithelial dysplasia (OED) is a pre-malignant histopathological diagnosis given to lesions of the oral cavity. Predicting OED grade or whether a case will transition to malignancy is critical for early detection and appropriate treatment. OED typically begins in the lower third of the epithelium before progressing upwards with grade severity, thus we have suggested that segmenting intra-epithelial layers, in addition to individual nuclei, may enable researchers to evaluate important layer-specific morphological features for grade/malignancy prediction. We present HoVer-Net+, a deep learning framework to simultaneously segment (and classify) nuclei and (intra-)epithelial layers in H&E stained slides from OED cases. The proposed architecture consists of an encoder branch and four decoder branches for simultaneous instance segmentation of nuclei and semantic segmentation of the epithelial layers. We show that the proposed model achieves the state-of-the-art (SOTA) performance in both tasks, with no additional costs when compared to previous SOTA methods for each task. To the best of our knowledge, ours is the first method for simultaneous nuclear instance segmentation and semantic tissue segmentation, with potential for use in computational pathology for other similar simultaneous tasks and for future studies into malignancy prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge