Syed Ali Khurram
Whole Slide Image Classification of Salivary Gland Tumours
Aug 22, 2024

Abstract:This work shows promising results using multiple instance learning on salivary gland tumours in classifying cancers on whole slide images. Utilising CTransPath as a patch-level feature extractor and CLAM as a feature aggregator, an F1 score of over 0.88 and AUROC of 0.92 are obtained for detecting cancer in whole slide images.
An Attention Based Pipeline for Identifying Pre-Cancer Lesions in Head and Neck Clinical Images
May 07, 2024



Abstract:Early detection of cancer can help improve patient prognosis by early intervention. Head and neck cancer is diagnosed in specialist centres after a surgical biopsy, however, there is a potential for these to be missed leading to delayed diagnosis. To overcome these challenges, we present an attention based pipeline that identifies suspected lesions, segments, and classifies them as non-dysplastic, dysplastic and cancerous lesions. We propose (a) a vision transformer based Mask R-CNN network for lesion detection and segmentation of clinical images, and (b) Multiple Instance Learning (MIL) based scheme for classification. Current results show that the segmentation model produces segmentation masks and bounding boxes with up to 82% overlap accuracy score on unseen external test data and surpassing reviewed segmentation benchmarks. Next, a classification F1-score of 85% on the internal cohort test set. An app has been developed to perform lesion segmentation taken via a smart device. Future work involves employing endoscopic video data for precise early detection and prognosis.
An Automated Pipeline for Tumour-Infiltrating Lymphocyte Scoring in Breast Cancer
Nov 21, 2023Abstract:Tumour-infiltrating lymphocytes (TILs) are considered as a valuable prognostic markers in both triple-negative and human epidermal growth factor receptor 2 (HER2) positive breast cancer. In this study, we introduce an innovative deep learning pipeline based on the Efficient-UNet architecture to predict the TILs score for breast cancer whole-slide images (WSIs). We first segment tumour and stromal regions in order to compute a tumour bulk mask. We then detect TILs within the tumour-associated stroma, generating a TILs score by closely mirroring the pathologist's workflow. Our method exhibits state-of-the-art performance in segmenting tumour/stroma areas and TILs detection, as demonstrated by internal cross-validation on the TiGER Challenge training dataset and evaluation on the final leaderboards. Additionally, our TILs score proves competitive in predicting survival outcomes within the same challenge, underscoring the clinical relevance and potential of our automated TILs scoring pipeline as a breast cancer prognostic tool.
Transformer-based Model for Oral Epithelial Dysplasia Segmentation
Nov 09, 2023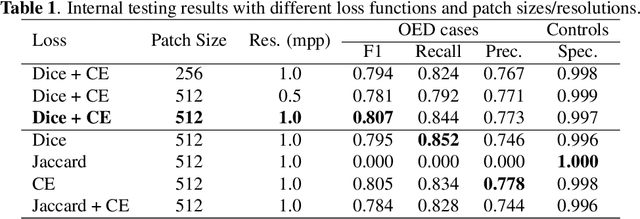
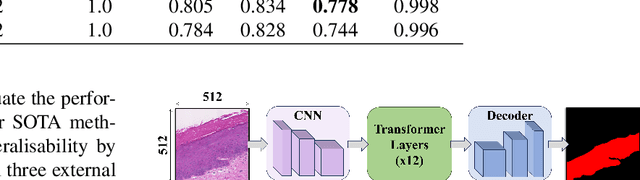
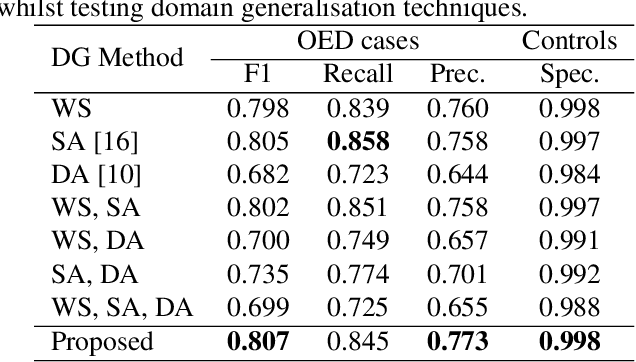
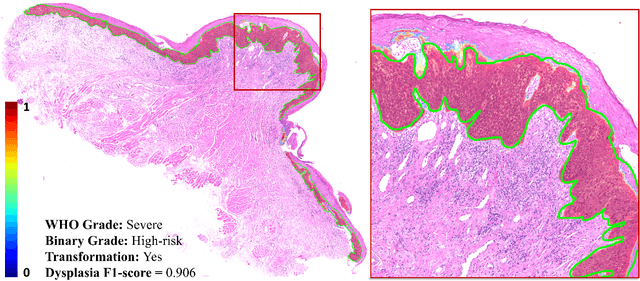
Abstract:Oral epithelial dysplasia (OED) is a premalignant histopathological diagnosis given to lesions of the oral cavity. OED grading is subject to large inter/intra-rater variability, resulting in the under/over-treatment of patients. We developed a new Transformer-based pipeline to improve detection and segmentation of OED in haematoxylin and eosin (H&E) stained whole slide images (WSIs). Our model was trained on OED cases (n = 260) and controls (n = 105) collected using three different scanners, and validated on test data from three external centres in the United Kingdom and Brazil (n = 78). Our internal experiments yield a mean F1-score of 0.81 for OED segmentation, which reduced slightly to 0.71 on external testing, showing good generalisability, and gaining state-of-the-art results. This is the first externally validated study to use Transformers for segmentation in precancerous histology images. Our publicly available model shows great promise to be the first step of a fully-integrated pipeline, allowing earlier and more efficient OED diagnosis, ultimately benefiting patient outcomes.
A Fully Automated and Explainable Algorithm for the Prediction of Malignant Transformation in Oral Epithelial Dysplasia
Jul 06, 2023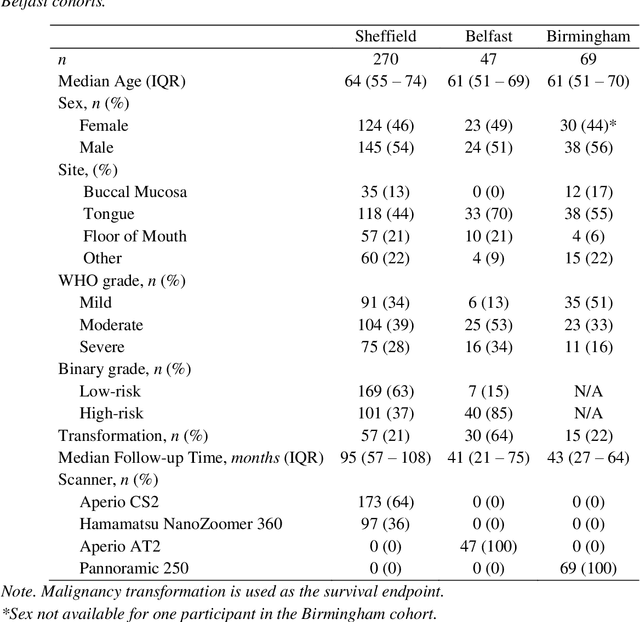
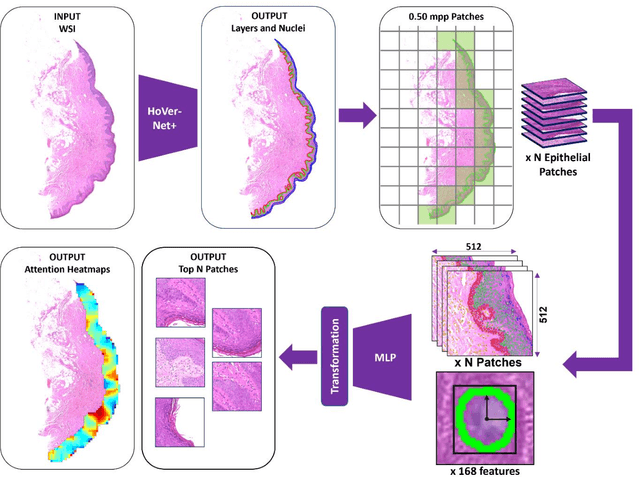
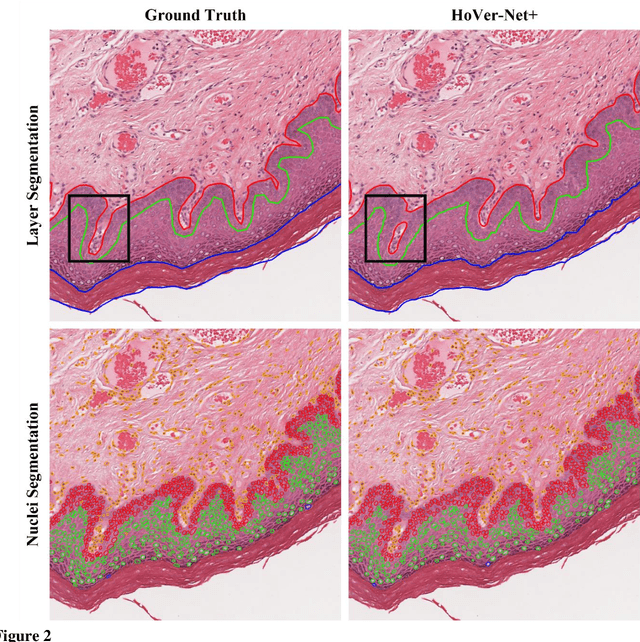
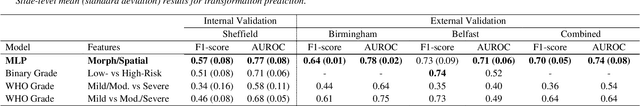
Abstract:Oral epithelial dysplasia (OED) is a premalignant histopathological diagnosis given to lesions of the oral cavity. Its grading suffers from significant inter-/intra- observer variability, and does not reliably predict malignancy progression, potentially leading to suboptimal treatment decisions. To address this, we developed a novel artificial intelligence algorithm that can assign an Oral Malignant Transformation (OMT) risk score, based on histological patterns in the in Haematoxylin and Eosin stained whole slide images, to quantify the risk of OED progression. The algorithm is based on the detection and segmentation of nuclei within (and around) the epithelium using an in-house segmentation model. We then employed a shallow neural network fed with interpretable morphological/spatial features, emulating histological markers. We conducted internal cross-validation on our development cohort (Sheffield; n = 193 cases) followed by independent validation on two external cohorts (Birmingham and Belfast; n = 92 cases). The proposed OMTscore yields an AUROC = 0.74 in predicting whether an OED progresses to malignancy or not. Survival analyses showed the prognostic value of our OMTscore for predicting malignancy transformation, when compared to the manually-assigned WHO and binary grades. Analysis of the correctly predicted cases elucidated the presence of peri-epithelial and epithelium-infiltrating lymphocytes in the most predictive patches of cases that transformed (p < 0.0001). This is the first study to propose a completely automated algorithm for predicting OED transformation based on interpretable nuclear features, whilst being validated on external datasets. The algorithm shows better-than-human-level performance for prediction of OED malignant transformation and offers a promising solution to the challenges of grading OED in routine clinical practice.
LYSTO: The Lymphocyte Assessment Hackathon and Benchmark Dataset
Jan 16, 2023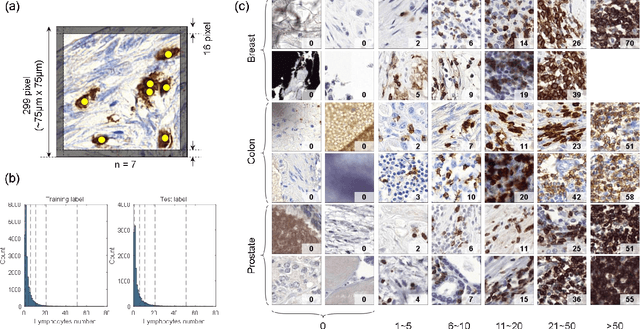
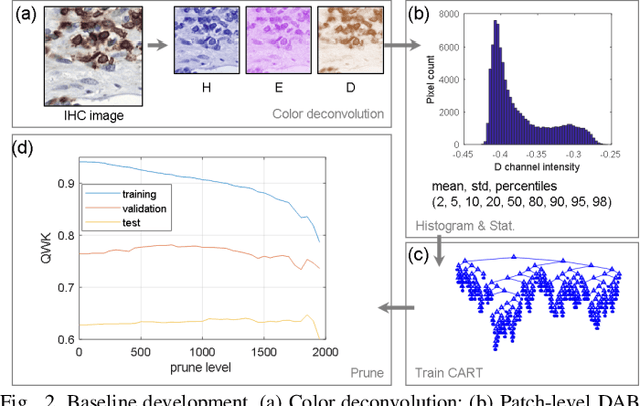
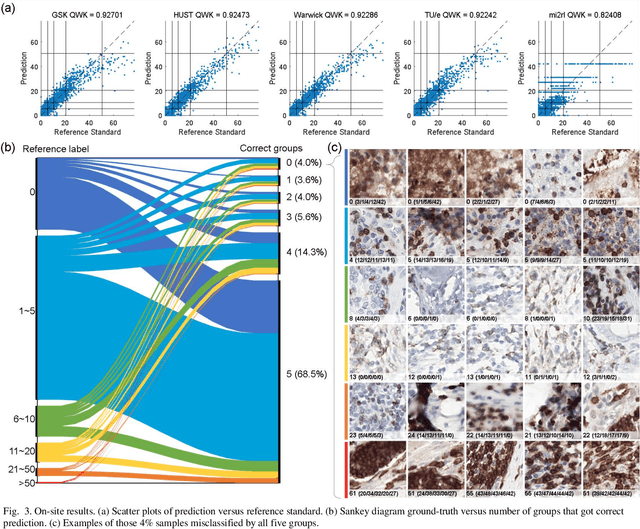
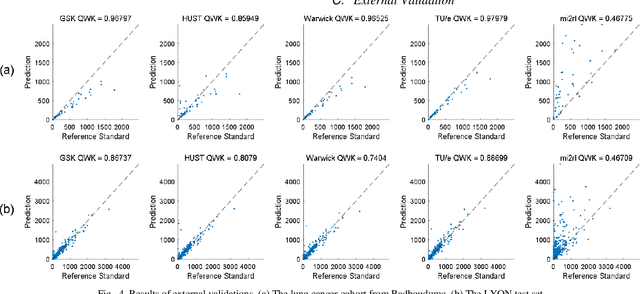
Abstract:We introduce LYSTO, the Lymphocyte Assessment Hackathon, which was held in conjunction with the MICCAI 2019 Conference in Shenzen (China). The competition required participants to automatically assess the number of lymphocytes, in particular T-cells, in histopathological images of colon, breast, and prostate cancer stained with CD3 and CD8 immunohistochemistry. Differently from other challenges setup in medical image analysis, LYSTO participants were solely given a few hours to address this problem. In this paper, we describe the goal and the multi-phase organization of the hackathon; we describe the proposed methods and the on-site results. Additionally, we present post-competition results where we show how the presented methods perform on an independent set of lung cancer slides, which was not part of the initial competition, as well as a comparison on lymphocyte assessment between presented methods and a panel of pathologists. We show that some of the participants were capable to achieve pathologist-level performance at lymphocyte assessment. After the hackathon, LYSTO was left as a lightweight plug-and-play benchmark dataset on grand-challenge website, together with an automatic evaluation platform. LYSTO has supported a number of research in lymphocyte assessment in oncology. LYSTO will be a long-lasting educational challenge for deep learning and digital pathology, it is available at https://lysto.grand-challenge.org/.
TIAger: Tumor-Infiltrating Lymphocyte Scoring in Breast Cancer for the TiGER Challenge
Jun 23, 2022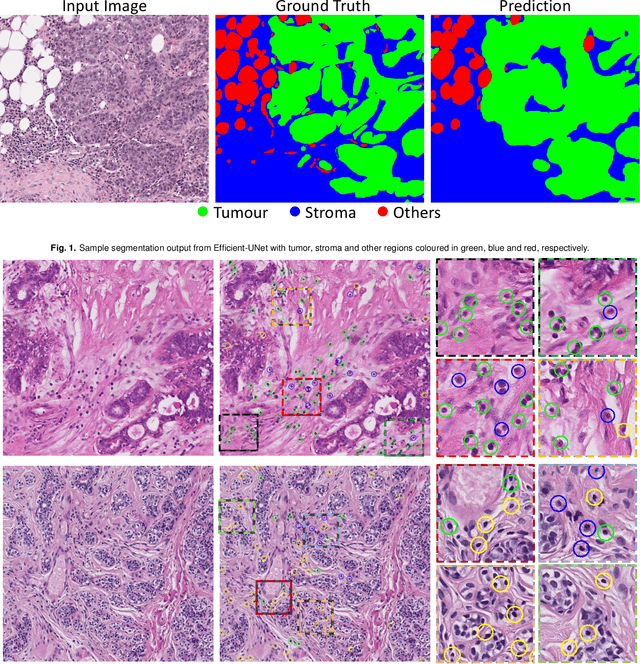
Abstract:The quantification of tumor-infiltrating lymphocytes (TILs) has been shown to be an independent predictor for prognosis of breast cancer patients. Typically, pathologists give an estimate of the proportion of the stromal region that contains TILs to obtain a TILs score. The Tumor InfiltratinG lymphocytes in breast cancER (TiGER) challenge, aims to assess the prognostic significance of computer-generated TILs scores for predicting survival as part of a Cox proportional hazards model. For this challenge, as the TIAger team, we have developed an algorithm to first segment tumor vs. stroma, before localising the tumor bulk region for TILs detection. Finally, we use these outputs to generate a TILs score for each case. On preliminary testing, our approach achieved a tumor-stroma weighted Dice score of 0.791 and a FROC score of 0.572 for lymphocytic detection. For predicting survival, our model achieved a C-index of 0.719. These results achieved first place across the preliminary testing leaderboards of the TiGER challenge.
Rank the triplets: A ranking-based multiple instance learning framework for detecting HPV infection in head and neck cancers using routine H&E images
Jun 16, 2022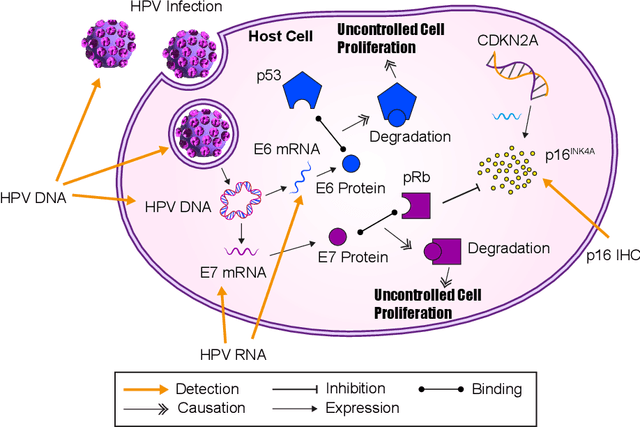

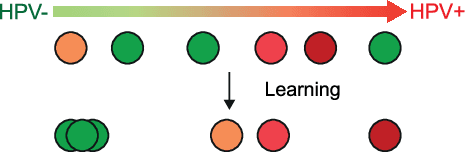
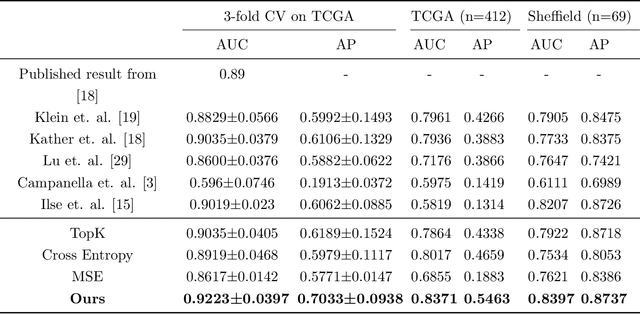
Abstract:The aetiology of head and neck squamous cell carcinoma (HNSCC) involves multiple carcinogens such as alcohol, tobacco and infection with human papillomavirus (HPV). As the HPV infection influences the prognosis, treatment and survival of patients with HNSCC, it is important to determine the HPV status of these tumours. In this paper, we propose a novel triplet-ranking loss function and a multiple instance learning pipeline for HPV status prediction. This achieves a new state-of-the-art performance in HPV detection using only the routine H&E stained WSIs on two HNSCC cohorts. Furthermore, a comprehensive tumour microenvironment profiling was performed, which characterised the unique patterns between HPV+/- HNSCC from genomic, immunology and cellular perspectives. Positive correlations of the proposed score with different subtypes of T cells (e.g. T cells follicular helper, CD8+ T cells), and negative correlations with macrophages and connective cells (e.g. fibroblast) were identified, which is in line with clinical findings. Unique gene expression profiles were also identified with respect to HPV infection status, and is in line with existing findings.
On Smart Gaze based Annotation of Histopathology Images for Training of Deep Convolutional Neural Networks
Feb 06, 2022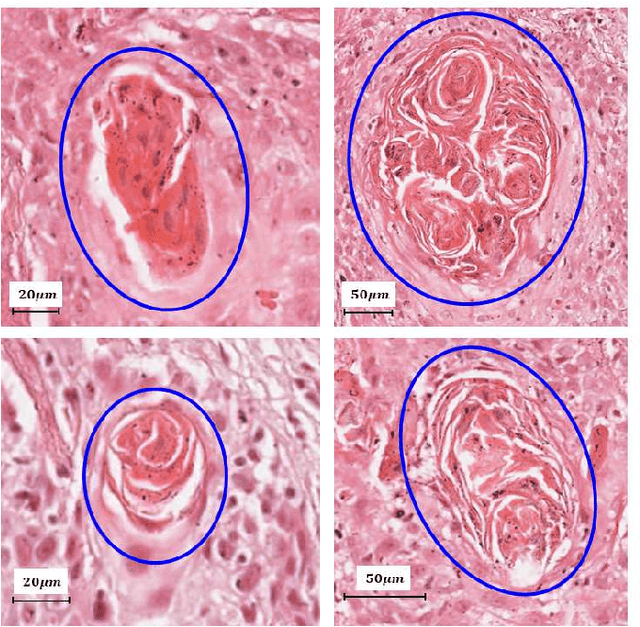
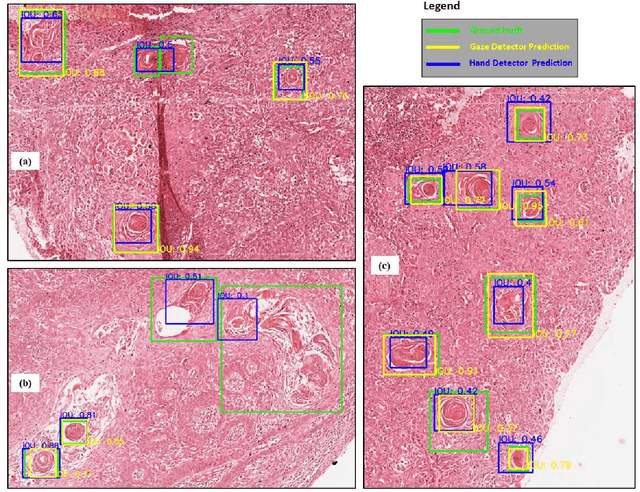
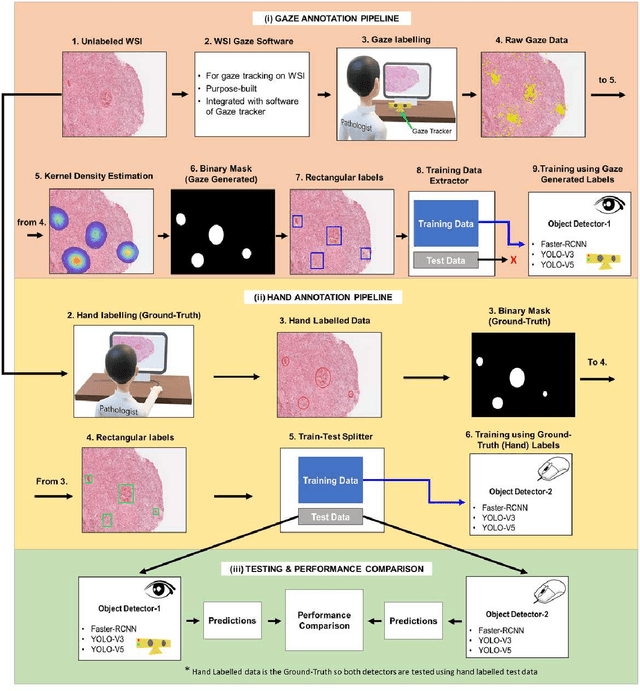
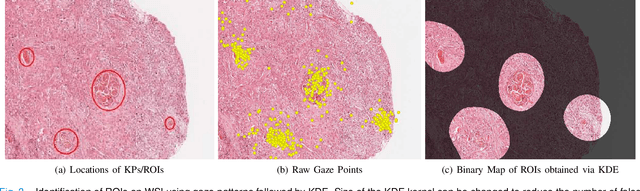
Abstract:Unavailability of large training datasets is a bottleneck that needs to be overcome to realize the true potential of deep learning in histopathology applications. Although slide digitization via whole slide imaging scanners has increased the speed of data acquisition, labeling of virtual slides requires a substantial time investment from pathologists. Eye gaze annotations have the potential to speed up the slide labeling process. This work explores the viability and timing comparisons of eye gaze labeling compared to conventional manual labeling for training object detectors. Challenges associated with gaze based labeling and methods to refine the coarse data annotations for subsequent object detection are also discussed. Results demonstrate that gaze tracking based labeling can save valuable pathologist time and delivers good performance when employed for training a deep object detector. Using the task of localization of Keratin Pearls in cases of oral squamous cell carcinoma as a test case, we compare the performance gap between deep object detectors trained using hand-labelled and gaze-labelled data. On average, compared to `Bounding-box' based hand-labeling, gaze-labeling required $57.6\%$ less time per label and compared to `Freehand' labeling, gaze-labeling required on average $85\%$ less time per label.
Stain-Robust Mitotic Figure Detection for the Mitosis Domain Generalization Challenge
Sep 29, 2021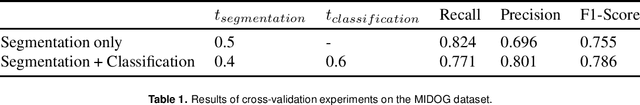
Abstract:The detection of mitotic figures from different scanners/sites remains an important topic of research, owing to its potential in assisting clinicians with tumour grading. The MItosis DOmain Generalization (MIDOG) challenge aims to test the robustness of detection models on unseen data from multiple scanners for this task. We present a short summary of the approach employed by the TIA Centre team to address this challenge. Our approach is based on a hybrid detection model, where mitotic candidates are segmented on stain normalised images, before being refined by a deep learning classifier. Cross-validation on the training images achieved the F1-score of 0.786 and 0.765 on the preliminary test set, demonstrating the generalizability of our model to unseen data from new scanners.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge