Shan E Ahmed Raza
MultiSurv: A Multimodal Deep Survival Framework for Prostrate and Bladder Cancer
Sep 05, 2025Abstract:Accurate prediction of time-to-event outcomes is a central challenge in oncology, with significant implications for treatment planning and patient management. In this work, we present MultiSurv, a multimodal deep survival model utilising DeepHit with a projection layer and inter-modality cross-attention, which integrates heterogeneous patient data, including clinical, MRI, RNA-seq and whole-slide pathology features. The model is designed to capture complementary prognostic signals across modalities and estimate individualised time-to-biochemical recurrence in prostate cancer and time-to-cancer recurrence in bladder cancer. Our approach was evaluated in the context of the CHIMERA Grand Challenge, across two of the three provided tasks. For Task 1 (prostate cancer bio-chemical recurrence prediction), the proposed framework achieved a concordance index (C-index) of 0.843 on 5-folds cross-validation and 0.818 on CHIMERA development set, demonstrating robust discriminatory ability. For Task 3 (bladder cancer recurrence prediction), the model obtained a C-index of 0.662 on 5-folds cross-validation and 0.457 on development set, highlighting its adaptability and potential for clinical translation. These results suggest that leveraging multimodal integration with deep survival learning provides a promising pathway toward personalised risk stratification in prostate and bladder cancer. Beyond the challenge setting, our framework is broadly applicable to survival prediction tasks involving heterogeneous biomedical data.
From Traditional to Deep Learning Approaches in Whole Slide Image Registration: A Methodological Review
Feb 26, 2025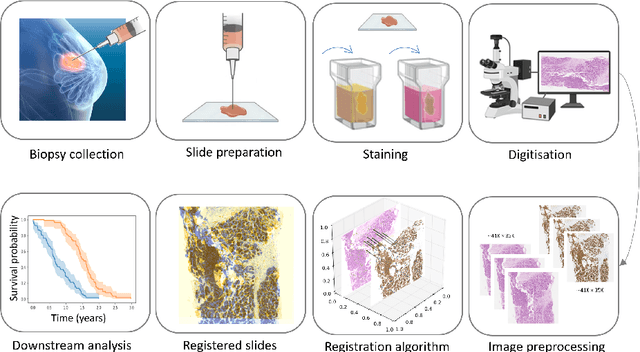
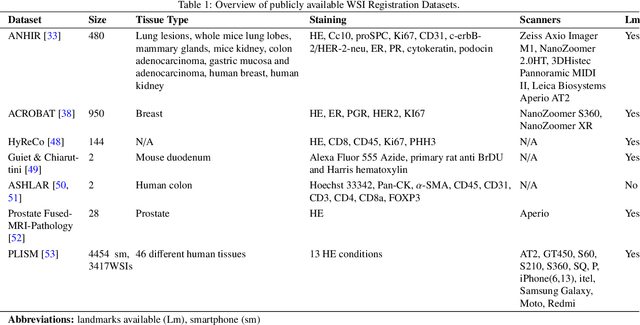
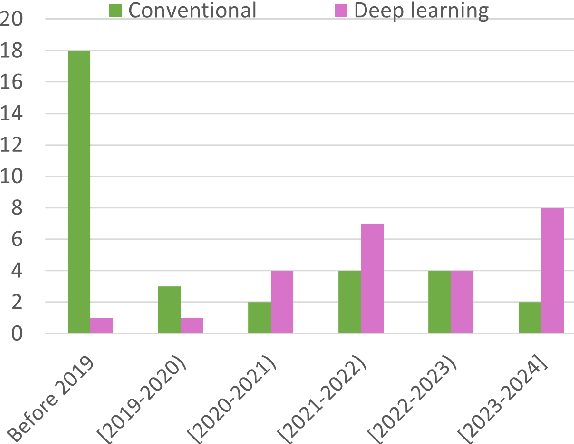
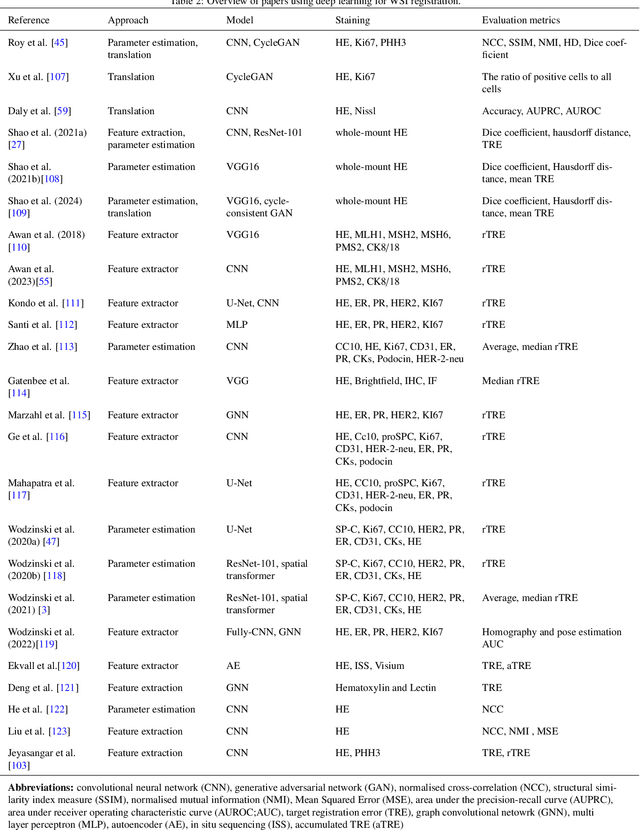
Abstract:Whole slide image (WSI) registration is an essential task for analysing the tumour microenvironment (TME) in histopathology. It involves the alignment of spatial information between WSIs of the same section or serial sections of a tissue sample. The tissue sections are usually stained with single or multiple biomarkers before imaging, and the goal is to identify neighbouring nuclei along the Z-axis for creating a 3D image or identifying subclasses of cells in the TME. This task is considerably more challenging compared to radiology image registration, such as magnetic resonance imaging or computed tomography, due to various factors. These include gigapixel size of images, variations in appearance between differently stained tissues, changes in structure and morphology between non-consecutive sections, and the presence of artefacts, tears, and deformations. Currently, there is a noticeable gap in the literature regarding a review of the current approaches and their limitations, as well as the challenges and opportunities they present. We aim to provide a comprehensive understanding of the available approaches and their application for various purposes. Furthermore, we investigate current deep learning methods used for WSI registration, emphasising their diverse methodologies. We examine the available datasets and explore tools and software employed in the field. Finally, we identify open challenges and potential future trends in this area of research.
Gland Segmentation Using SAM With Cancer Grade as a Prompt
Jan 27, 2025Abstract:Cancer grade is a critical clinical criterion that can be used to determine the degree of cancer malignancy. Revealing the condition of the glands, a precise gland segmentation can assist in a more effective cancer grade classification. In machine learning, binary classification information about glands (i.e., benign and malignant) can be utilized as a prompt for gland segmentation and cancer grade classification. By incorporating prior knowledge of the benign or malignant classification of the gland, the model can anticipate the likely appearance of the target, leading to better segmentation performance. We utilize Segment Anything Model to solve the segmentation task, by taking advantage of its prompt function and applying appropriate modifications to the model structure and training strategies. We improve the results from fine-tuned Segment Anything Model and produce SOTA results using this approach.
Deep Learning Based Segmentation of Blood Vessels from H&E Stained Oesophageal Adenocarcinoma Whole-Slide Images
Jan 21, 2025Abstract:Blood vessels (BVs) play a critical role in the Tumor Micro-Environment (TME), potentially influencing cancer progression and treatment response. However, manually quantifying BVs in Hematoxylin and Eosin (H&E) stained images is challenging and labor-intensive due to their heterogeneous appearances. We propose a novel approach of constructing guiding maps to improve the performance of state-of-the-art segmentation models for BV segmentation, the guiding maps encourage the models to learn representative features of BVs. This is particularly beneficial for computational pathology, where labeled training data is often limited and large models are prone to overfitting. We have quantitative and qualitative results to demonstrate the efficacy of our approach in improving segmentation accuracy. In future, we plan to validate this method to segment BVs across various tissue types and investigate the role of cellular structures in relation to BVs in the TME.
CellOMaps: A Compact Representation for Robust Classification of Lung Adenocarcinoma Growth Patterns
Jan 14, 2025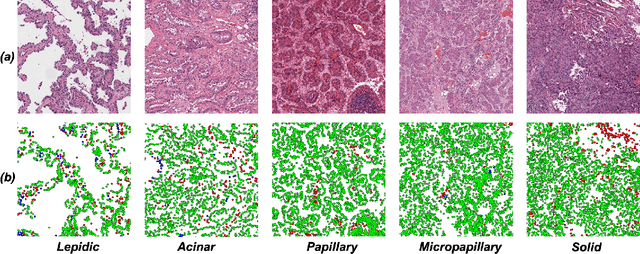



Abstract:Lung adenocarcinoma (LUAD) is a morphologically heterogeneous disease, characterized by five primary histological growth patterns. The classification of such patterns is crucial due to their direct relation to prognosis but the high subjectivity and observer variability pose a major challenge. Although several studies have developed machine learning methods for growth pattern classification, they either only report the predominant pattern per slide or lack proper evaluation. We propose a generalizable machine learning pipeline capable of classifying lung tissue into one of the five patterns or as non-tumor. The proposed pipeline's strength lies in a novel compact Cell Organization Maps (cellOMaps) representation that captures the cellular spatial patterns from Hematoxylin and Eosin whole slide images (WSIs). The proposed pipeline provides state-of-the-art performance on LUAD growth pattern classification when evaluated on both internal unseen slides and external datasets, significantly outperforming the current approaches. In addition, our preliminary results show that the model's outputs can be used to predict patients Tumor Mutational Burden (TMB) levels.
An Attention Based Pipeline for Identifying Pre-Cancer Lesions in Head and Neck Clinical Images
May 07, 2024



Abstract:Early detection of cancer can help improve patient prognosis by early intervention. Head and neck cancer is diagnosed in specialist centres after a surgical biopsy, however, there is a potential for these to be missed leading to delayed diagnosis. To overcome these challenges, we present an attention based pipeline that identifies suspected lesions, segments, and classifies them as non-dysplastic, dysplastic and cancerous lesions. We propose (a) a vision transformer based Mask R-CNN network for lesion detection and segmentation of clinical images, and (b) Multiple Instance Learning (MIL) based scheme for classification. Current results show that the segmentation model produces segmentation masks and bounding boxes with up to 82% overlap accuracy score on unseen external test data and surpassing reviewed segmentation benchmarks. Next, a classification F1-score of 85% on the internal cohort test set. An app has been developed to perform lesion segmentation taken via a smart device. Future work involves employing endoscopic video data for precise early detection and prognosis.
Nuclei-Location Based Point Set Registration of Multi-Stained Whole Slide Images
Apr 25, 2024



Abstract:Whole Slide Images (WSIs) provide exceptional detail for studying tissue architecture at the cell level. To study tumour microenvironment (TME) with the context of various protein biomarkers and cell sub-types, analysis and registration of features using multi-stained WSIs is often required. Multi-stained WSI pairs normally suffer from rigid and non-rigid deformities in addition to slide artefacts and control tissue which present challenges at precise registration. Traditional registration methods mainly focus on global rigid/non-rigid registration but struggle with aligning slides with complex tissue deformations at the nuclei level. However, nuclei level non-rigid registration is essential for downstream tasks such as cell sub-type analysis in the context of protein biomarker signatures. This paper focuses on local level non-rigid registration using a nuclei-location based point set registration approach for aligning multi-stained WSIs. We exploit the spatial distribution of nuclei that is prominent and consistent (to a large level) across different stains to establish a spatial correspondence. We evaluate our approach using the HYRECO dataset consisting of 54 re-stained images of H\&E and PHH3 image pairs. The approach can be extended to other IHC and IF stained WSIs considering a good nuclei detection algorithm is accessible. The performance of the model is tested against established registration algorithms and is shown to outperform the model for nuclei level registration.
An AI based Digital Score of Tumour-Immune Microenvironment Predicts Benefit to Maintenance Immunotherapy in Advanced Oesophagogastric Adenocarcinoma
Feb 29, 2024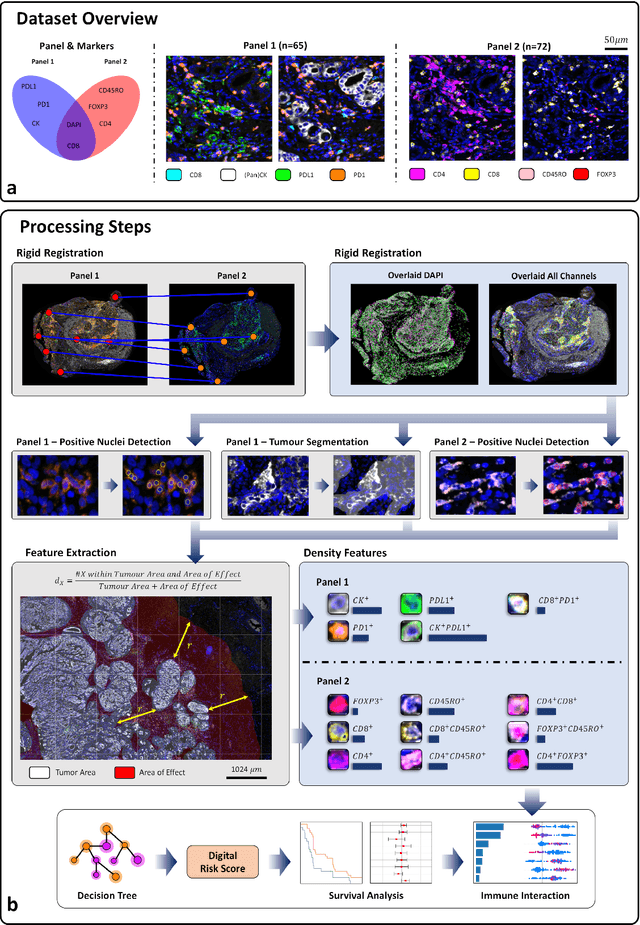
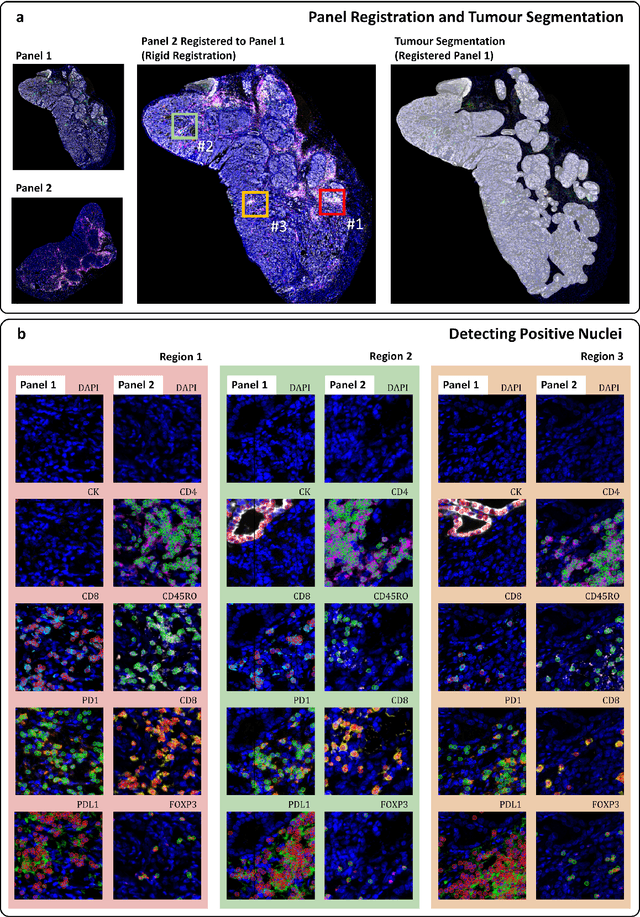
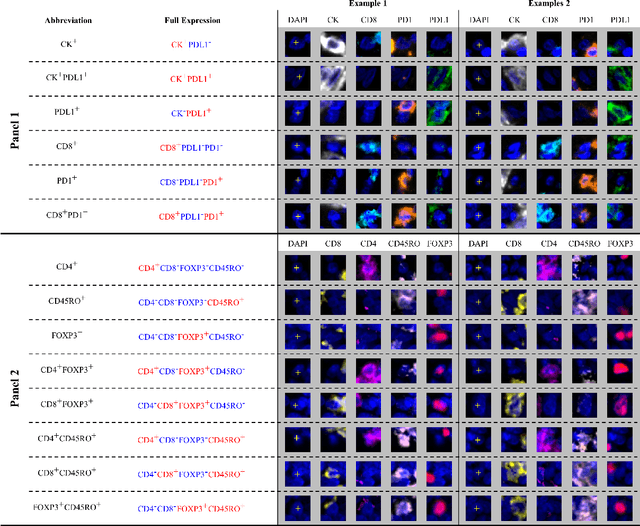
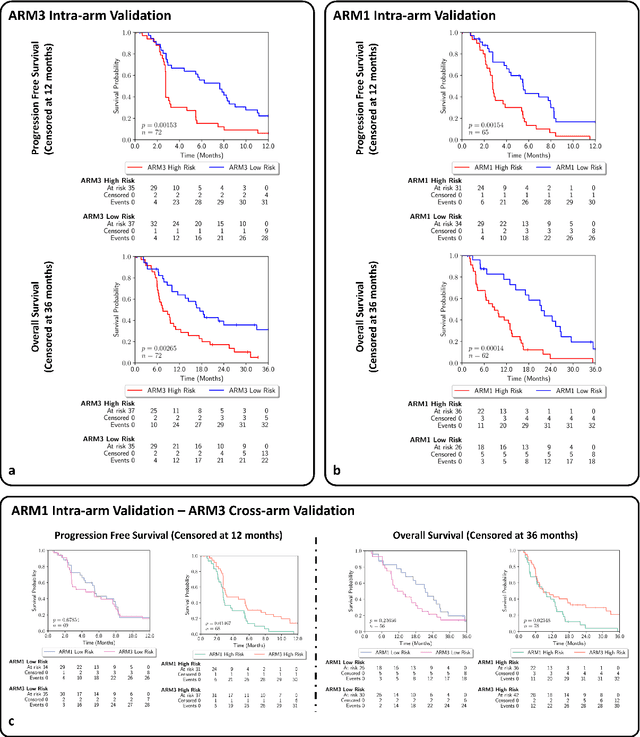
Abstract:Gastric and oesophageal (OG) cancers are the leading causes of cancer mortality worldwide. In OG cancers, recent studies have showed that PDL1 immune checkpoint inhibitors (ICI) in combination with chemotherapy improves patient survival. However, our understanding of the tumour immune microenvironment in OG cancers remains limited. In this study, we interrogate multiplex immunofluorescence (mIF) images taken from patients with advanced Oesophagogastric Adenocarcinoma (OGA) who received first-line fluoropyrimidine and platinum-based chemotherapy in the PLATFORM trial (NCT02678182) to predict the efficacy of the treatment and to explore the biological basis of patients responding to maintenance durvalumab (PDL1 inhibitor). Our proposed Artificial Intelligence (AI) based marker successfully identified responder from non-responder (p < 0.05) as well as those who could potentially benefit from ICI with statistical significance (p < 0.05) for both progression free and overall survival. Our findings suggest that T cells that express FOXP3 seem to heavily influence the patient treatment response and survival outcome. We also observed that higher levels of CD8+PD1+ cells are consistently linked to poor prognosis for both OS and PFS, regardless of ICI.
Cell Maps Representation For Lung Adenocarcinoma Growth Patterns Classification In Whole Slide Images
Nov 27, 2023


Abstract:Lung adenocarcinoma is a morphologically heterogeneous disease, characterized by five primary histologic growth patterns. The quantity of these patterns can be related to tumor behavior and has a significant impact on patient prognosis. In this work, we propose a novel machine learning pipeline capable of classifying tissue tiles into one of the five patterns or as non-tumor, with an Area Under the Receiver Operating Characteristic Curve (AUCROC) score of 0.97. Our model's strength lies in its comprehensive consideration of cellular spatial patterns, where it first generates cell maps from Hematoxylin and Eosin (H&E) whole slide images (WSIs), which are then fed into a convolutional neural network classification model. Exploiting these cell maps provides the model with robust generalizability to new data, achieving approximately 30% higher accuracy on unseen test-sets compared to current state of the art approaches. The insights derived from our model can be used to predict prognosis, enhancing patient outcomes.
An Automated Pipeline for Tumour-Infiltrating Lymphocyte Scoring in Breast Cancer
Nov 21, 2023Abstract:Tumour-infiltrating lymphocytes (TILs) are considered as a valuable prognostic markers in both triple-negative and human epidermal growth factor receptor 2 (HER2) positive breast cancer. In this study, we introduce an innovative deep learning pipeline based on the Efficient-UNet architecture to predict the TILs score for breast cancer whole-slide images (WSIs). We first segment tumour and stromal regions in order to compute a tumour bulk mask. We then detect TILs within the tumour-associated stroma, generating a TILs score by closely mirroring the pathologist's workflow. Our method exhibits state-of-the-art performance in segmenting tumour/stroma areas and TILs detection, as demonstrated by internal cross-validation on the TiGER Challenge training dataset and evaluation on the final leaderboards. Additionally, our TILs score proves competitive in predicting survival outcomes within the same challenge, underscoring the clinical relevance and potential of our automated TILs scoring pipeline as a breast cancer prognostic tool.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge