Neda Zamani Tajeddin
Stain-Robust Mitotic Figure Detection for the Mitosis Domain Generalization Challenge
Sep 29, 2021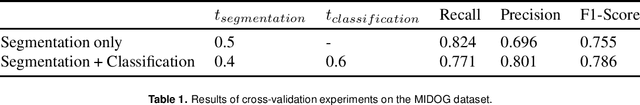
Abstract:The detection of mitotic figures from different scanners/sites remains an important topic of research, owing to its potential in assisting clinicians with tumour grading. The MItosis DOmain Generalization (MIDOG) challenge aims to test the robustness of detection models on unseen data from multiple scanners for this task. We present a short summary of the approach employed by the TIA Centre team to address this challenge. Our approach is based on a hybrid detection model, where mitotic candidates are segmented on stain normalised images, before being refined by a deep learning classifier. Cross-validation on the training images achieved the F1-score of 0.786 and 0.765 on the preliminary test set, demonstrating the generalizability of our model to unseen data from new scanners.
Robust Interactive Semantic Segmentation of Pathology Images with Minimal User Input
Aug 30, 2021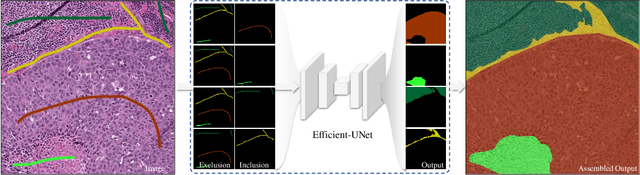
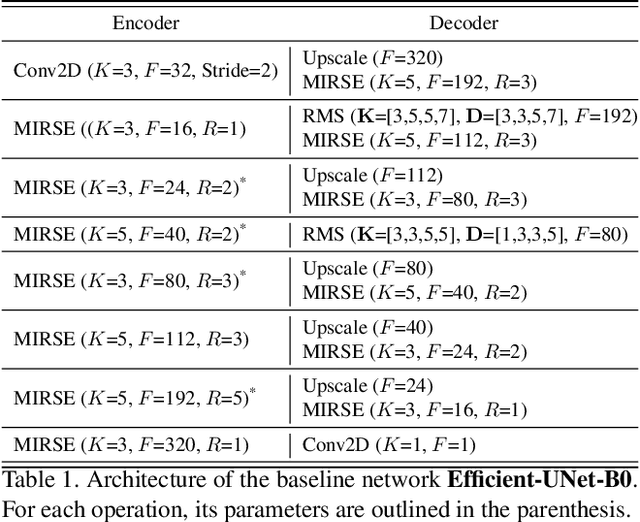
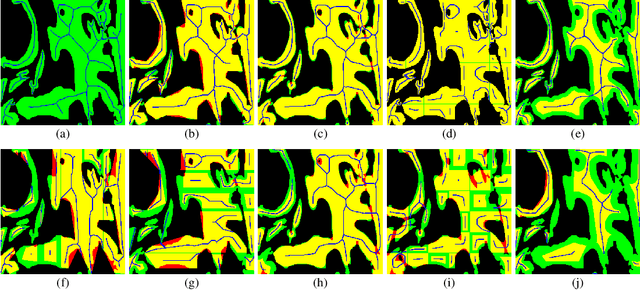
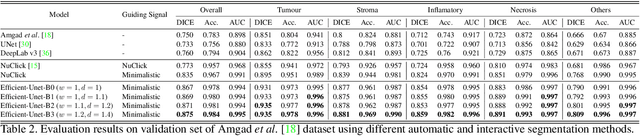
Abstract:From the simple measurement of tissue attributes in pathology workflow to designing an explainable diagnostic/prognostic AI tool, access to accurate semantic segmentation of tissue regions in histology images is a prerequisite. However, delineating different tissue regions manually is a laborious, time-consuming and costly task that requires expert knowledge. On the other hand, the state-of-the-art automatic deep learning models for semantic segmentation require lots of annotated training data and there are only a limited number of tissue region annotated images publicly available. To obviate this issue in computational pathology projects and collect large-scale region annotations efficiently, we propose an efficient interactive segmentation network that requires minimum input from the user to accurately annotate different tissue types in the histology image. The user is only required to draw a simple squiggle inside each region of interest so it will be used as the guiding signal for the model. To deal with the complex appearance and amorph geometry of different tissue regions we introduce several automatic and minimalistic guiding signal generation techniques that help the model to become robust against the variation in the user input. By experimenting on a dataset of breast cancer images, we show that not only does our proposed method speed up the interactive annotation process, it can also outperform the existing automatic and interactive region segmentation models.
Automatic Recognition of the Supraspinatus Tendinopathy from Ultrasound Images using Convolutional Neural Networks
Nov 23, 2020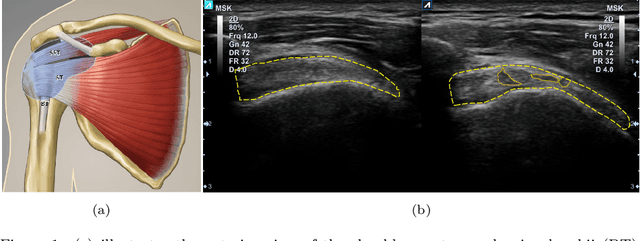

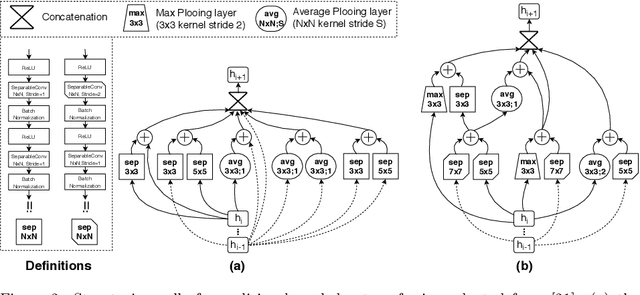
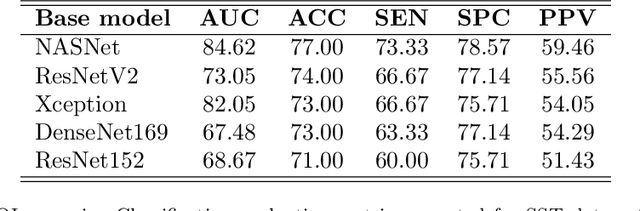
Abstract:Tendon injuries like tendinopathies, full and partial thickness tears are prevalent, and the supraspinatus tendon (SST) is the most vulnerable ones in the rotator cuff. Early diagnosis of SST tendinopathies is of high importance and hard to achieve using ultrasound imaging. In this paper, an automatic tendinopathy recognition framework based on convolutional neural networks has been proposed to assist the diagnosis. This framework has two essential parts of tendon segmentation and classification. Tendon segmentation is done through a novel network, NASUNet, which follows an encoder-decoder architecture paradigm and utilizes a multi-scale Enlarging cell. Moreover, a general classification pipeline has been proposed for tendinopathy recognition, which supports different base models as the feature extractor engine. Two feature maps comprising positional information of the tendon region have been introduced as the network input to make the classification network spatial-aware. To evaluate the tendinopathy recognition system, a data set consisting of 100 SST ultrasound images have been acquired, in which tendinopathy cases are double-verified by magnetic resonance imaging. In both segmentation and classification tasks, lack of training data has been compensated by incorporating knowledge transferring, transfer learning, and data augmentation techniques. In cross-validation experiments, the proposed tendinopathy recognition model achieves 91% accuracy, 86.67% sensitivity, and 92.86% specificity, showing state-of-the-art performance against other models.
Leveraging Transfer Learning for Segmenting Lesions and their Attributes in Dermoscopy Images
Sep 23, 2018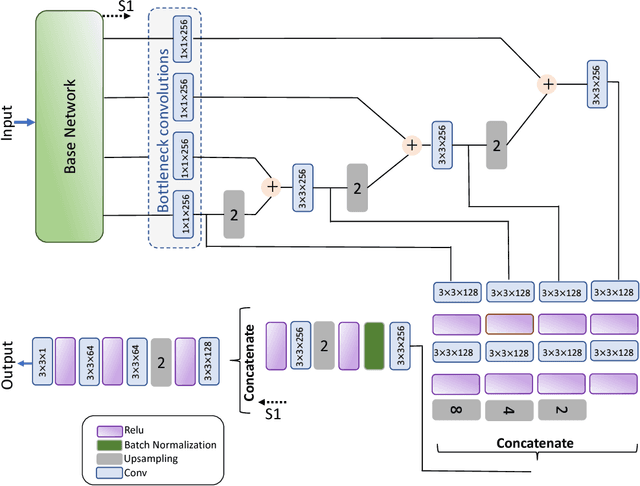

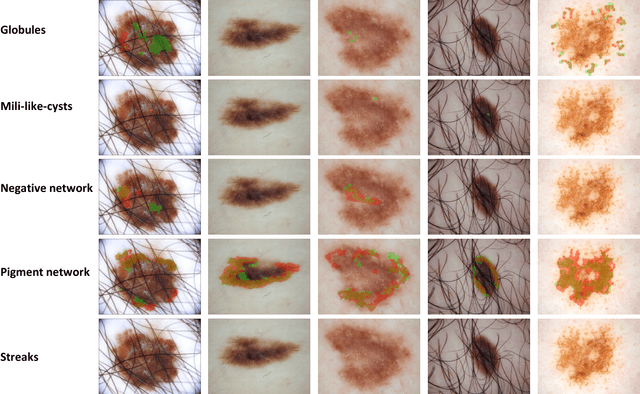
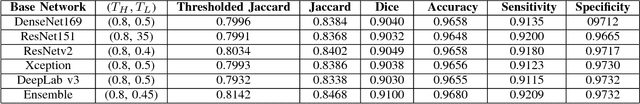
Abstract:Computer-aided diagnosis systems for classification of different type of skin lesions have been an active field of research in recent decades. It has been shown that introducing lesions and their attributes masks into lesion classification pipeline can greatly improve the performance. In this paper, we propose a framework by incorporating transfer learning for segmenting lesions and their attributes based on the convolutional neural networks. The proposed framework is inspired by the well-known UNet architecture. It utilizes a variety of pre-trained networks in the encoding path and generates the prediction map by combining multi-scale information in decoding path using a pyramid pooling manner. To circumvent the lack of training data and increase the proposed model generalization, an extensive set of novel augmentation routines have been applied during the training of the network. Moreover, for each task of lesion and attribute segmentation, a specific loss function has been designed to obviate the training phase difficulties. Finally, the prediction for each task is generated by ensembling the outputs from different models. The proposed approach achieves promising results on the cross-validation experiments on the ISIC2018- Task1 and Task2 data sets.
Supervised Saliency Map Driven Segmentation of the Lesions in Dermoscopic Images
Jun 07, 2018
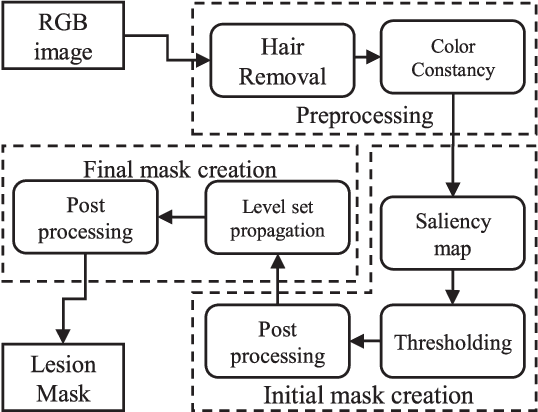


Abstract:Lesion segmentation is the first step in most automatic melanoma recognition systems. Deficiencies and difficulties in dermoscopic images such as color inconstancy, hair occlusion, dark corners and color charts make lesion segmentation an intricate task. In order to detect the lesion in the presence of these problems, we propose a supervised saliency detection method tailored for dermoscopic images based on the discriminative regional feature integration (DRFI). DRFI method incorporates multi-level segmentation, regional contrast, property, background descriptors, and a random forest regressor to create saliency scores for each region in the image. In our improved saliency detection method, mDRFI, we have added some new features to regional property descriptors. Also, in order to achieve more robust regional background descriptors, a thresholding algorithm is proposed to obtain a new pseudo-background region. Findings reveal that mDRFI is superior to DRFI in detecting the lesion as the salient object in dermoscopic images. The proposed overall lesion segmentation framework uses detected saliency map to construct an initial mask of the lesion through thresholding and post-processing operations. The initial mask is then evolving in a level set framework to fit better on the lesion's boundaries. The results of evaluation tests on three public datasets show that our proposed segmentation method outperforms the other conventional state-of-the-art segmentation algorithms and its performance is comparable with most recent approaches that are based on deep convolutional neural networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge