Navid Alemi Koohbanani
LYSTO: The Lymphocyte Assessment Hackathon and Benchmark Dataset
Jan 16, 2023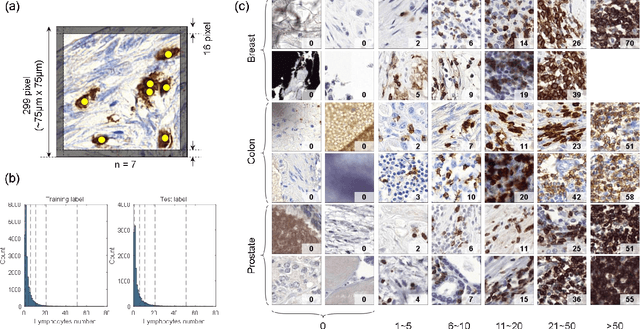
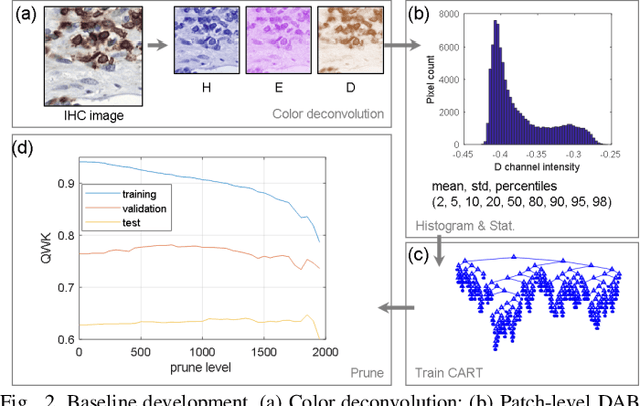
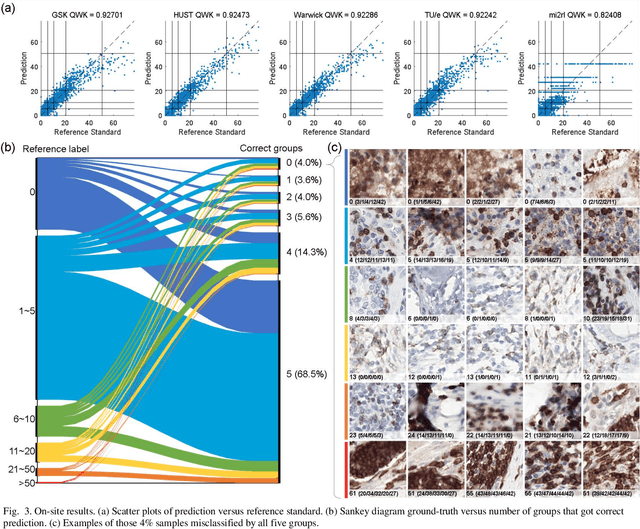
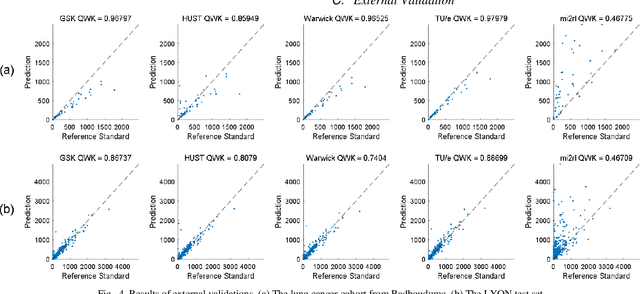
Abstract:We introduce LYSTO, the Lymphocyte Assessment Hackathon, which was held in conjunction with the MICCAI 2019 Conference in Shenzen (China). The competition required participants to automatically assess the number of lymphocytes, in particular T-cells, in histopathological images of colon, breast, and prostate cancer stained with CD3 and CD8 immunohistochemistry. Differently from other challenges setup in medical image analysis, LYSTO participants were solely given a few hours to address this problem. In this paper, we describe the goal and the multi-phase organization of the hackathon; we describe the proposed methods and the on-site results. Additionally, we present post-competition results where we show how the presented methods perform on an independent set of lung cancer slides, which was not part of the initial competition, as well as a comparison on lymphocyte assessment between presented methods and a panel of pathologists. We show that some of the participants were capable to achieve pathologist-level performance at lymphocyte assessment. After the hackathon, LYSTO was left as a lightweight plug-and-play benchmark dataset on grand-challenge website, together with an automatic evaluation platform. LYSTO has supported a number of research in lymphocyte assessment in oncology. LYSTO will be a long-lasting educational challenge for deep learning and digital pathology, it is available at https://lysto.grand-challenge.org/.
Robust Interactive Semantic Segmentation of Pathology Images with Minimal User Input
Aug 30, 2021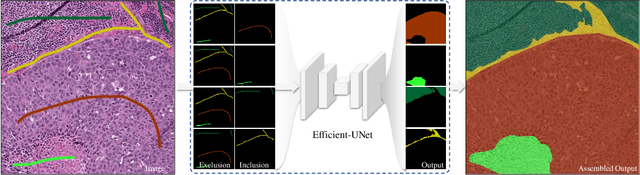
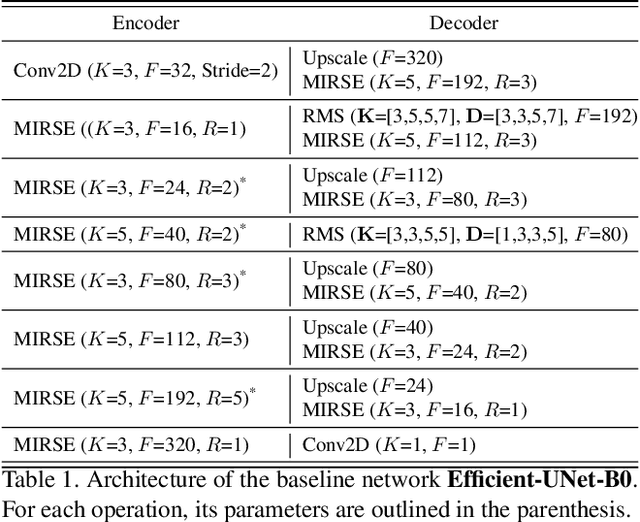
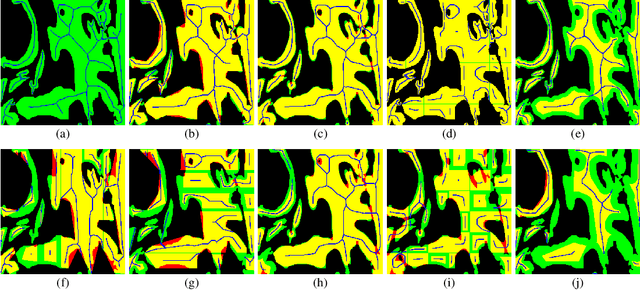
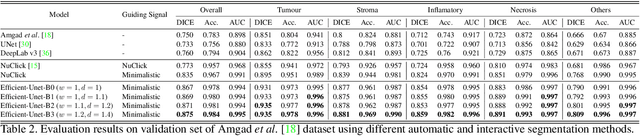
Abstract:From the simple measurement of tissue attributes in pathology workflow to designing an explainable diagnostic/prognostic AI tool, access to accurate semantic segmentation of tissue regions in histology images is a prerequisite. However, delineating different tissue regions manually is a laborious, time-consuming and costly task that requires expert knowledge. On the other hand, the state-of-the-art automatic deep learning models for semantic segmentation require lots of annotated training data and there are only a limited number of tissue region annotated images publicly available. To obviate this issue in computational pathology projects and collect large-scale region annotations efficiently, we propose an efficient interactive segmentation network that requires minimum input from the user to accurately annotate different tissue types in the histology image. The user is only required to draw a simple squiggle inside each region of interest so it will be used as the guiding signal for the model. To deal with the complex appearance and amorph geometry of different tissue regions we introduce several automatic and minimalistic guiding signal generation techniques that help the model to become robust against the variation in the user input. By experimenting on a dataset of breast cancer images, we show that not only does our proposed method speed up the interactive annotation process, it can also outperform the existing automatic and interactive region segmentation models.
Self-Path: Self-supervision for Classification of Pathology Images with Limited Annotations
Aug 12, 2020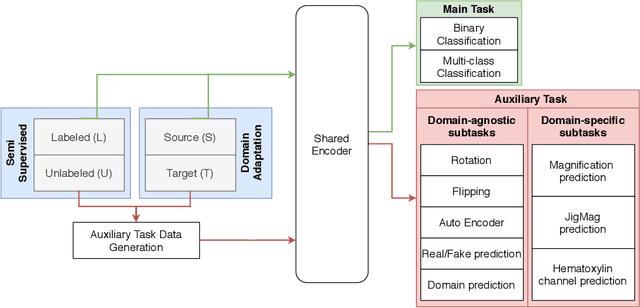
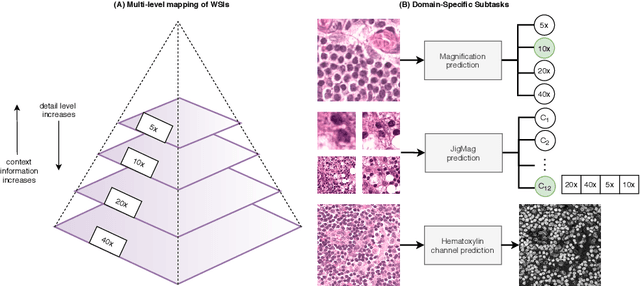
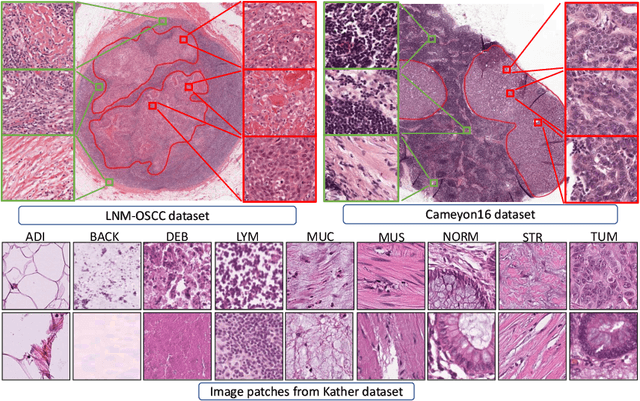
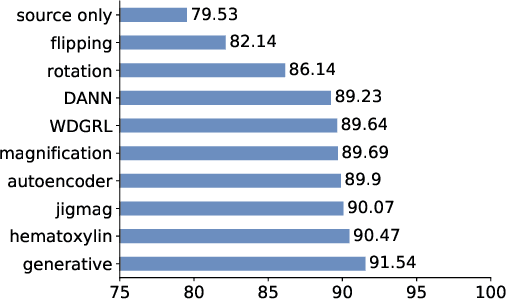
Abstract:While high-resolution pathology images lend themselves well to `data hungry' deep learning algorithms, obtaining exhaustive annotations on these images is a major challenge. In this paper, we propose a self-supervised CNN approach to leverage unlabeled data for learning generalizable and domain invariant representations in pathology images. The proposed approach, which we term as Self-Path, is a multi-task learning approach where the main task is tissue classification and pretext tasks are a variety of self-supervised tasks with labels inherent to the input data. We introduce novel domain specific self-supervision tasks that leverage contextual, multi-resolution and semantic features in pathology images for semi-supervised learning and domain adaptation. We investigate the effectiveness of Self-Path on 3 different pathology datasets. Our results show that Self-Path with the domain-specific pretext tasks achieves state-of-the-art performance for semi-supervised learning when small amounts of labeled data are available. Further, we show that Self-Path improves domain adaptation for classification of histology image patches when there is no labeled data available for the target domain. This approach can potentially be employed for other applications in computational pathology, where annotation budget is often limited or large amount of unlabeled image data is available.
NuClick: A Deep Learning Framework for Interactive Segmentation of Microscopy Images
May 29, 2020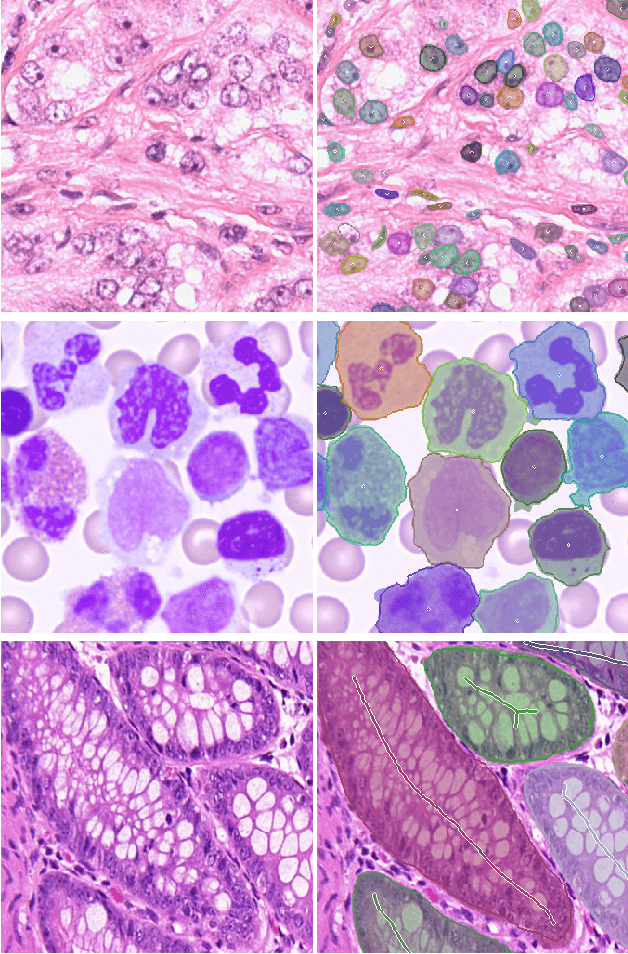
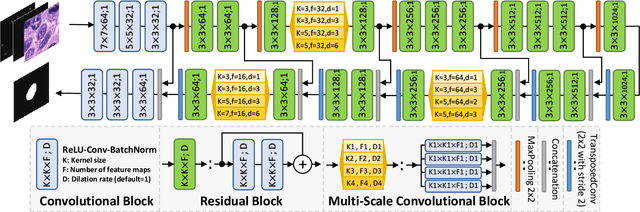
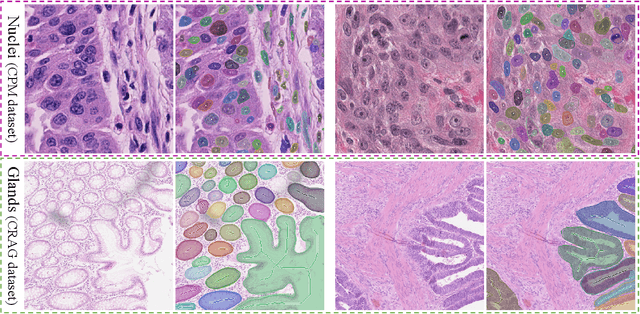
Abstract:Object Segmentation is an important step in the work-flow of computational pathology. Deep learning based models as the best forming models require huge amount of labeled data for precise and reliable prediction. However, collecting labeled data is expensive, because it necessarily involves expert knowledge. This is perhaps best illustrated by medical tasks where measurements call for expensive machinery and labels are the fruit of a time-consuming analysis that draws from multiple human experts. As nuclei, cells and glands are fundamental objects for downstream analysis in histology, in this paper we propose a simple CNN-based approach to speed up collecting segmentation annotation for these objects by utilizing minimum input from an annotator. We show for nuclei and cells as small objects, one click inside objects is enough to have precise annotation. For glands as large objects, providing a squiggle to show the extend of gland can guide the model to outline the exact boundaries. This supervisory signals are fed to network as an auxiliary channels along with RGB channels. With detailed experiments, we show that our approach is generalizable, robust against variations in the user input and that it can be used to obtain annotations for completely different domains. Practically, a model trained on the masks generated by NuClick could achieve first rank in LYON19 challenge. Furthermore, as the output of our framework, we release two data-sets: 1) a dataset of lymphocyte annotations within IHC images and 2) a dataset of WBCs annotated in blood sample images.
PanNuke Dataset Extension, Insights and Baselines
Apr 22, 2020
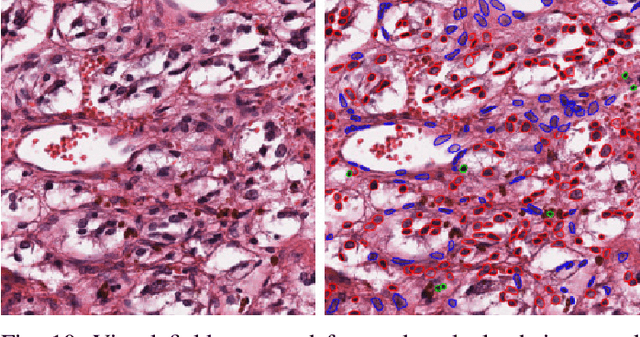
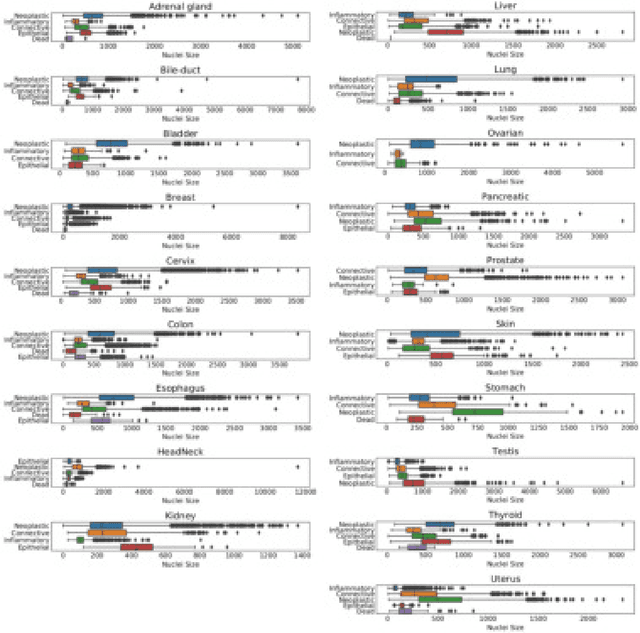
Abstract:The emerging area of computational pathology (CPath) is ripe ground for the application of deep learning (DL) methods to healthcare due to the sheer volume of raw pixel data in whole-slide images (WSIs) of cancerous tissue slides. However, it is imperative for the DL algorithms relying on nuclei-level details to be able to cope with data from `the clinical wild', which tends to be quite challenging. We study, and extend recently released PanNuke dataset consisting of ~200,000 nuclei categorized into 5 clinically important classes for the challenging tasks of segmenting and classifying nuclei in WSIs. Previous pan-cancer datasets consisted of only up to 9 different tissues and up to 21,000 unlabeled nuclei and just over 24,000 labeled nuclei with segmentation masks. PanNuke consists of 19 different tissue types that have been semi-automatically annotated and quality controlled by clinical pathologists, leading to a dataset with statistics similar to the clinical wild and with minimal selection bias. We study the performance of segmentation and classification models when applied to the proposed dataset and demonstrate the application of models trained on PanNuke to whole-slide images. We provide comprehensive statistics about the dataset and outline recommendations and research directions to address the limitations of existing DL tools when applied to real-world CPath applications.
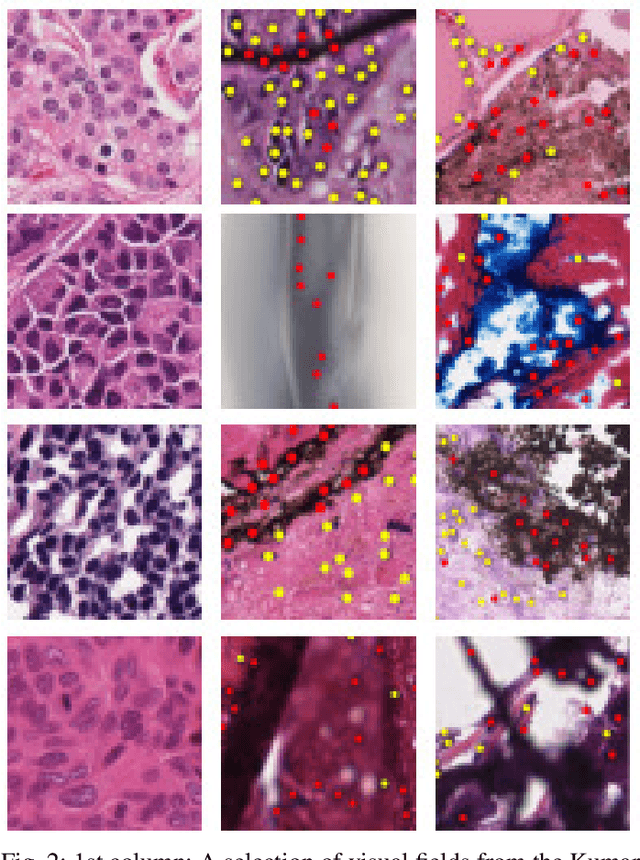
NuClick: From Clicks in the Nuclei to Nuclear Boundaries
Sep 07, 2019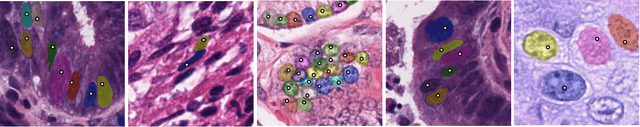
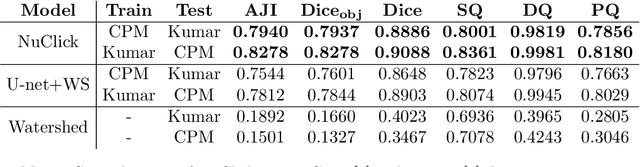
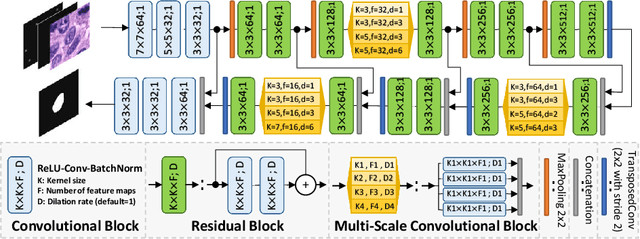
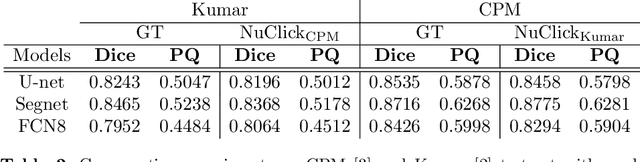
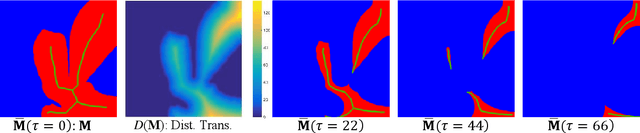
Abstract:Best performing nuclear segmentation methods are based on deep learning algorithms that require a large amount of annotated data. However, collecting annotations for nuclear segmentation is a very labor-intensive and time-consuming task. Thereby, providing a tool that can facilitate and speed up this procedure is very demanding. Here we propose a simple yet efficient framework based on convolutional neural networks, named NuClick, which can precisely segment nuclei boundaries by accepting a single point position (or click) inside each nucleus. Based on the clicked positions, inclusion and exclusion maps are generated which comprise 2D Gaussian distributions centered on those positions. These maps serve as guiding signals for the network as they are concatenated to the input image. The inclusion map focuses on the desired nucleus while the exclusion map indicates neighboring nuclei and improve the results of segmentation in scenes with nuclei clutter. The NuClick not only facilitates collecting more annotation from unseen data but also leads to superior segmentation output for deep models. It is also worth mentioning that an instance segmentation model trained on NuClick generated labels was able to rank first in LYON19 challenge.
CGC-Net: Cell Graph Convolutional Network for Grading of Colorectal Cancer Histology Images
Sep 03, 2019


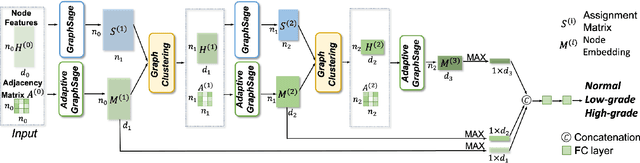
Abstract:Colorectal cancer (CRC) grading is typically carried out by assessing the degree of gland formation within histology images. To do this, it is important to consider the overall tissue micro-environment by assessing the cell-level information along with the morphology of the gland. However, current automated methods for CRC grading typically utilise small image patches and therefore fail to incorporate the entire tissue micro-architecture for grading purposes. To overcome the challenges of CRC grading, we present a novel cell-graph convolutional neural network (CGC-Net) that converts each large histology image into a graph, where each node is represented by a nucleus within the original image and cellular interactions are denoted as edges between these nodes according to node similarity. The CGC-Net utilises nuclear appearance features in addition to the spatial location of nodes to further boost the performance of the algorithm. To enable nodes to fuse multi-scale information, we introduce Adaptive GraphSage, which is a graph convolution technique that combines multi-level features in a data-driven way. Furthermore, to deal with redundancy in the graph, we propose a sampling technique that removes nodes in areas of dense nuclear activity. We show that modeling the image as a graph enables us to effectively consider a much larger image (around 16$\times$ larger) than traditional patch-based approaches and model the complex structure of the tissue micro-environment. We construct cell graphs with an average of over 3,000 nodes on a large CRC histology image dataset and report state-of-the-art results as compared to recent patch-based as well as contextual patch-based techniques, demonstrating the effectiveness of our method.
Nuclear Instance Segmentation using a Proposal-Free Spatially Aware Deep Learning Framework
Aug 27, 2019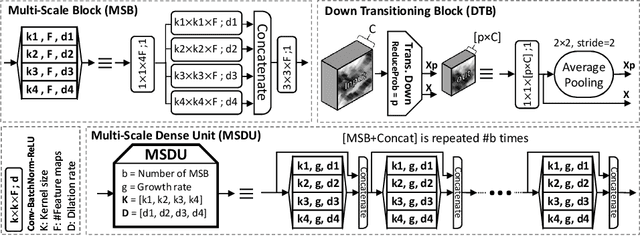
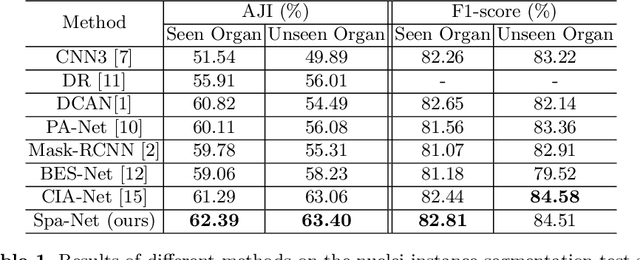
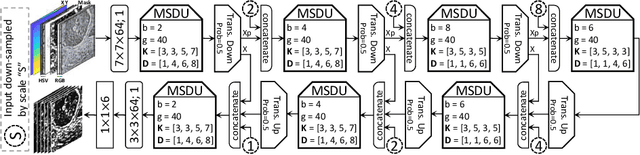
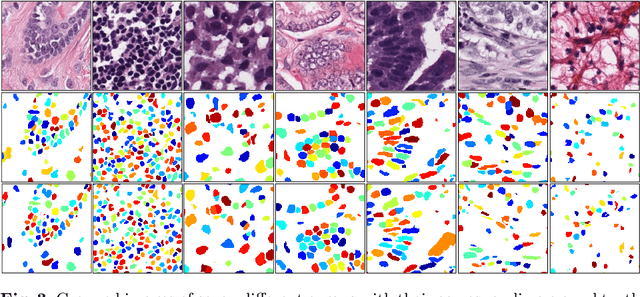
Abstract:Nuclear segmentation in histology images is a challenging task due to significant variations in the shape and appearance of nuclei. One of the main hurdles in nuclear instance segmentation is overlapping nuclei where a smart algorithm is needed to separate each nucleus. In this paper, we introduce a proposal-free deep learning based framework to address these challenges. To this end, we propose a spatially-aware network (SpaNet) to capture spatial information in a multi-scale manner. A dual-head variation of the SpaNet is first utilized to predict the pixel-wise segmentation and centroid detection maps of nuclei. Based on these outputs, a single-head SpaNet predicts the positional information related to each nucleus instance. Spectral clustering method is applied on the output of the last SpaNet, which utilizes the nuclear mask and the Gaussian-like detection map for determining the connected components and associated cluster identifiers, respectively. The output of the clustering method is the final nuclear instance segmentation mask. We applied our method on a publicly available multi-organ data set and achieved state-of-the-art performance for nuclear segmentation.
Methods for Segmentation and Classification of Digital Microscopy Tissue Images
Oct 31, 2018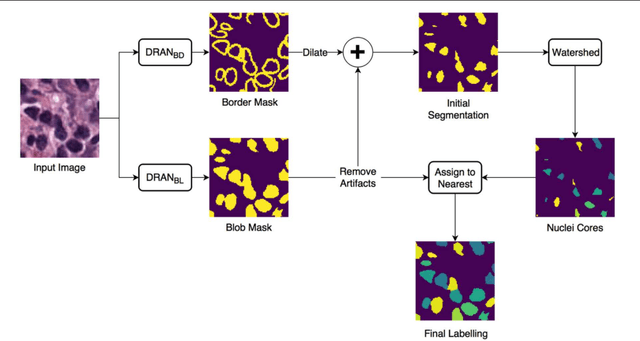

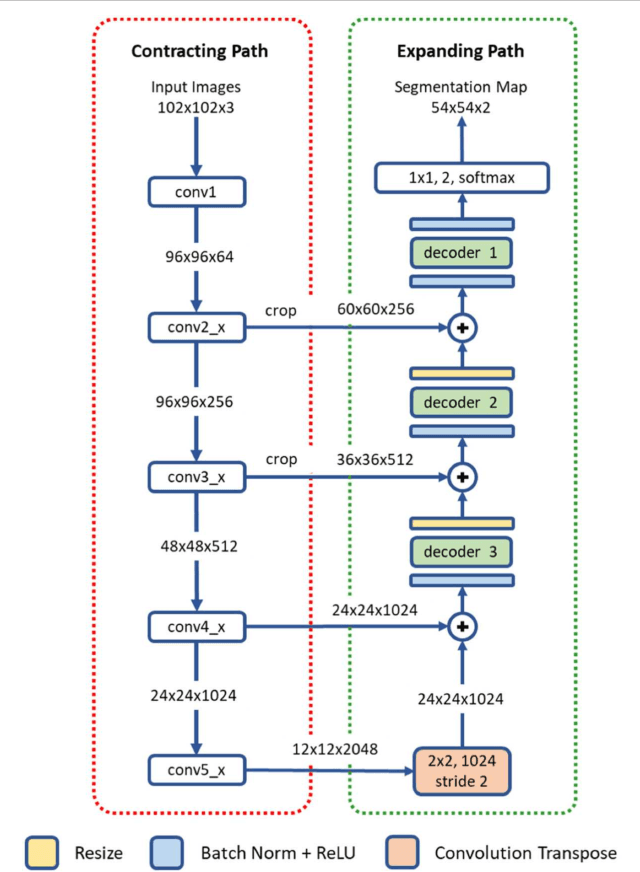
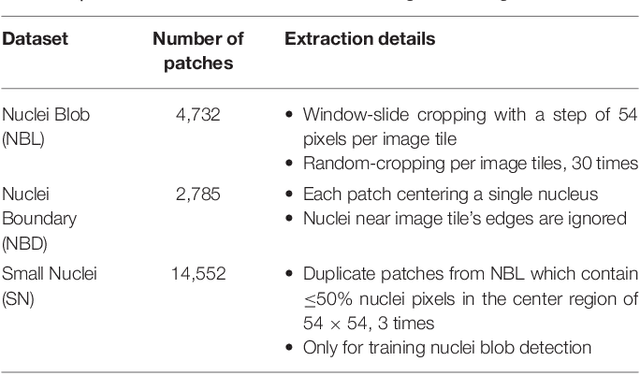
Abstract:High-resolution microscopy images of tissue specimens provide detailed information about the morphology of normal and diseased tissue. Image analysis of tissue morphology can help cancer researchers develop a better understanding of cancer biology. Segmentation of nuclei and classification of tissue images are two common tasks in tissue image analysis. Development of accurate and efficient algorithms for these tasks is a challenging problem because of the complexity of tissue morphology and tumor heterogeneity. In this paper we present two computer algorithms; one designed for segmentation of nuclei and the other for classification of whole slide tissue images. The segmentation algorithm implements a multiscale deep residual aggregation network to accurately segment nuclear material and then separate clumped nuclei into individual nuclei. The classification algorithm initially carries out patch-level classification via a deep learning method, then patch-level statistical and morphological features are used as input to a random forest regression model for whole slide image classification. The segmentation and classification algorithms were evaluated in the MICCAI 2017 Digital Pathology challenge. The segmentation algorithm achieved an accuracy score of 0.78. The classification algorithm achieved an accuracy score of 0.81.
Leveraging Transfer Learning for Segmenting Lesions and their Attributes in Dermoscopy Images
Sep 23, 2018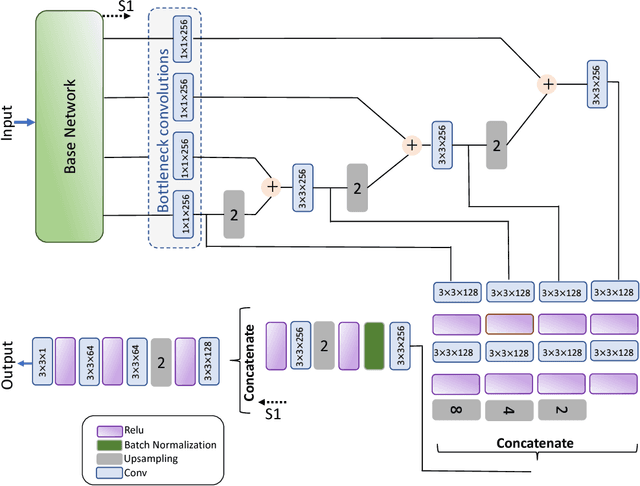

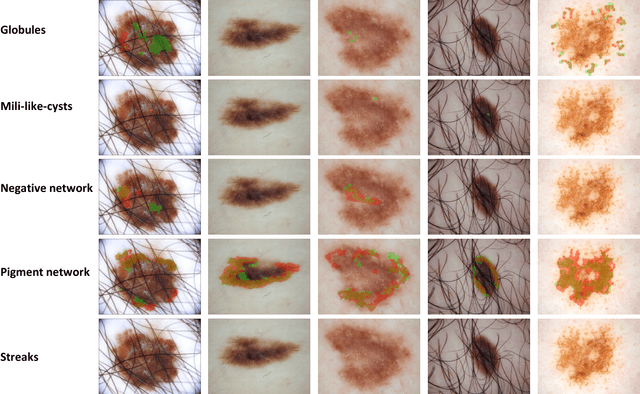
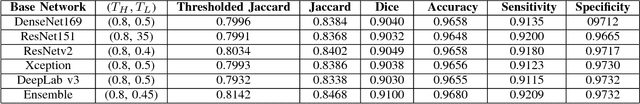
Abstract:Computer-aided diagnosis systems for classification of different type of skin lesions have been an active field of research in recent decades. It has been shown that introducing lesions and their attributes masks into lesion classification pipeline can greatly improve the performance. In this paper, we propose a framework by incorporating transfer learning for segmenting lesions and their attributes based on the convolutional neural networks. The proposed framework is inspired by the well-known UNet architecture. It utilizes a variety of pre-trained networks in the encoding path and generates the prediction map by combining multi-scale information in decoding path using a pyramid pooling manner. To circumvent the lack of training data and increase the proposed model generalization, an extensive set of novel augmentation routines have been applied during the training of the network. Moreover, for each task of lesion and attribute segmentation, a specific loss function has been designed to obviate the training phase difficulties. Finally, the prediction for each task is generated by ensembling the outputs from different models. The proposed approach achieves promising results on the cross-validation experiments on the ISIC2018- Task1 and Task2 data sets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge