Self-Path: Self-supervision for Classification of Pathology Images with Limited Annotations
Paper and Code
Aug 12, 2020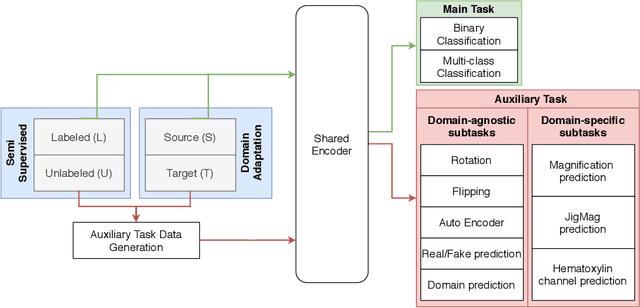
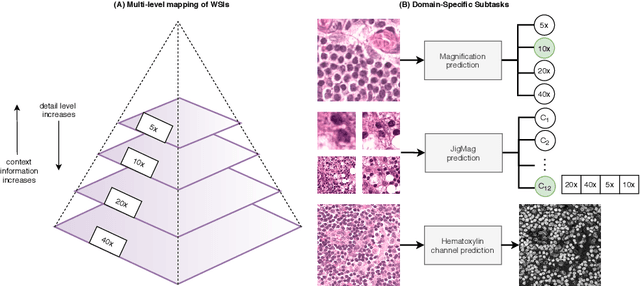
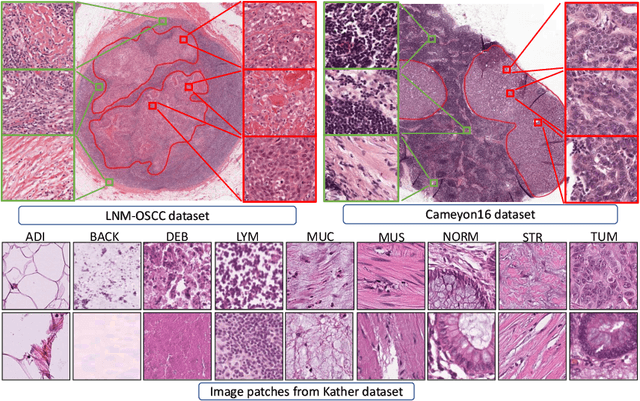
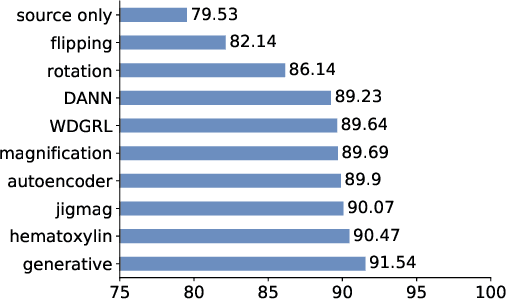
While high-resolution pathology images lend themselves well to `data hungry' deep learning algorithms, obtaining exhaustive annotations on these images is a major challenge. In this paper, we propose a self-supervised CNN approach to leverage unlabeled data for learning generalizable and domain invariant representations in pathology images. The proposed approach, which we term as Self-Path, is a multi-task learning approach where the main task is tissue classification and pretext tasks are a variety of self-supervised tasks with labels inherent to the input data. We introduce novel domain specific self-supervision tasks that leverage contextual, multi-resolution and semantic features in pathology images for semi-supervised learning and domain adaptation. We investigate the effectiveness of Self-Path on 3 different pathology datasets. Our results show that Self-Path with the domain-specific pretext tasks achieves state-of-the-art performance for semi-supervised learning when small amounts of labeled data are available. Further, we show that Self-Path improves domain adaptation for classification of histology image patches when there is no labeled data available for the target domain. This approach can potentially be employed for other applications in computational pathology, where annotation budget is often limited or large amount of unlabeled image data is available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge