Balagopal Unnikrishnan
Red Teaming Large Language Models for Healthcare
May 01, 2025Abstract:We present the design process and findings of the pre-conference workshop at the Machine Learning for Healthcare Conference (2024) entitled Red Teaming Large Language Models for Healthcare, which took place on August 15, 2024. Conference participants, comprising a mix of computational and clinical expertise, attempted to discover vulnerabilities -- realistic clinical prompts for which a large language model (LLM) outputs a response that could cause clinical harm. Red-teaming with clinicians enables the identification of LLM vulnerabilities that may not be recognised by LLM developers lacking clinical expertise. We report the vulnerabilities found, categorise them, and present the results of a replication study assessing the vulnerabilities across all LLMs provided.
Semi-supervised classification of radiology images with NoTeacher: A Teacher that is not Mean
Aug 10, 2021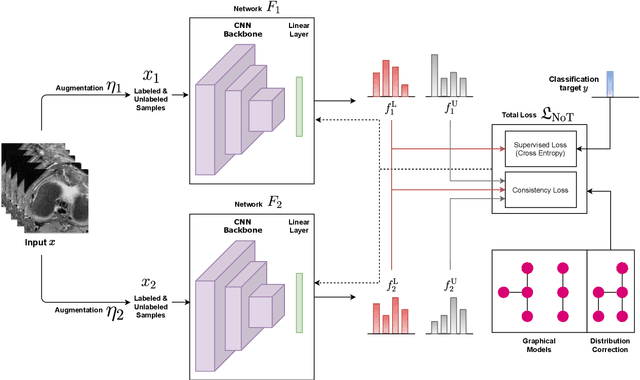
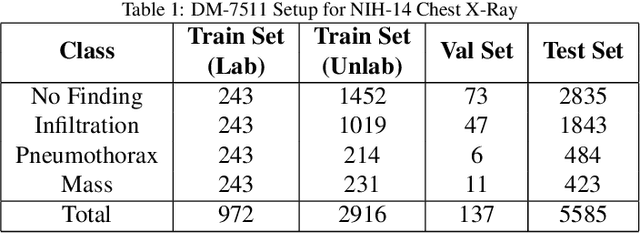
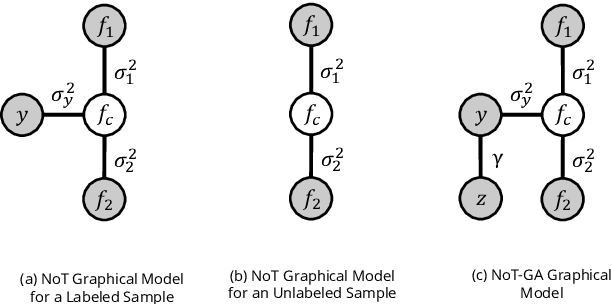
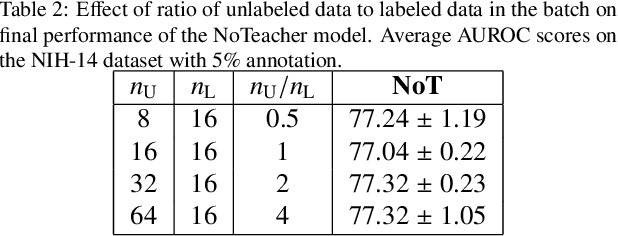
Abstract:Deep learning models achieve strong performance for radiology image classification, but their practical application is bottlenecked by the need for large labeled training datasets. Semi-supervised learning (SSL) approaches leverage small labeled datasets alongside larger unlabeled datasets and offer potential for reducing labeling cost. In this work, we introduce NoTeacher, a novel consistency-based SSL framework which incorporates probabilistic graphical models. Unlike Mean Teacher which maintains a teacher network updated via a temporal ensemble, NoTeacher employs two independent networks, thereby eliminating the need for a teacher network. We demonstrate how NoTeacher can be customized to handle a range of challenges in radiology image classification. Specifically, we describe adaptations for scenarios with 2D and 3D inputs, uni and multi-label classification, and class distribution mismatch between labeled and unlabeled portions of the training data. In realistic empirical evaluations on three public benchmark datasets spanning the workhorse modalities of radiology (X-Ray, CT, MRI), we show that NoTeacher achieves over 90-95% of the fully supervised AUROC with less than 5-15% labeling budget. Further, NoTeacher outperforms established SSL methods with minimal hyperparameter tuning, and has implications as a principled and practical option for semisupervised learning in radiology applications.
Self-Path: Self-supervision for Classification of Pathology Images with Limited Annotations
Aug 12, 2020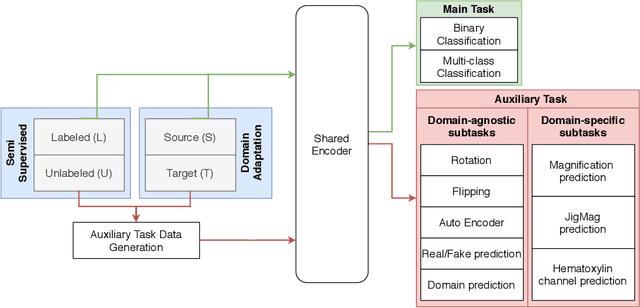
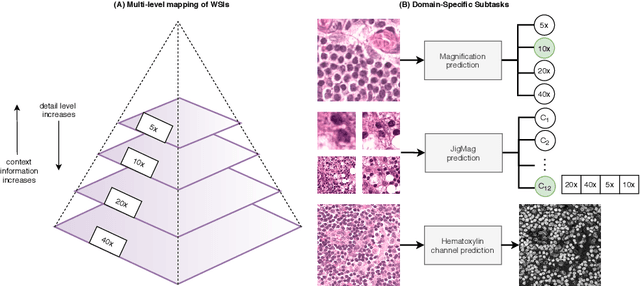
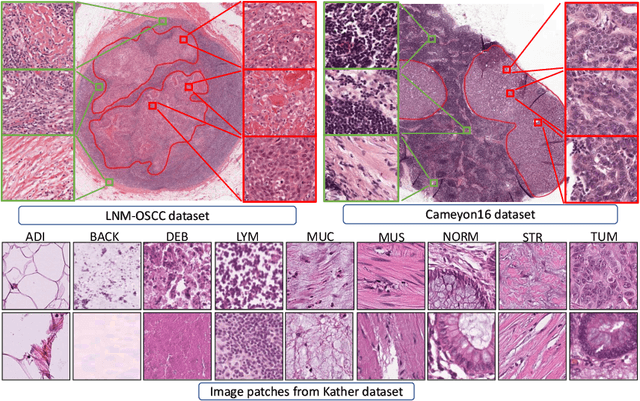
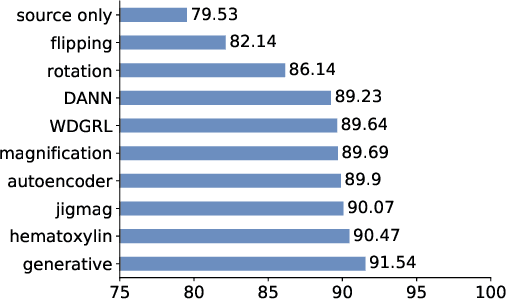
Abstract:While high-resolution pathology images lend themselves well to `data hungry' deep learning algorithms, obtaining exhaustive annotations on these images is a major challenge. In this paper, we propose a self-supervised CNN approach to leverage unlabeled data for learning generalizable and domain invariant representations in pathology images. The proposed approach, which we term as Self-Path, is a multi-task learning approach where the main task is tissue classification and pretext tasks are a variety of self-supervised tasks with labels inherent to the input data. We introduce novel domain specific self-supervision tasks that leverage contextual, multi-resolution and semantic features in pathology images for semi-supervised learning and domain adaptation. We investigate the effectiveness of Self-Path on 3 different pathology datasets. Our results show that Self-Path with the domain-specific pretext tasks achieves state-of-the-art performance for semi-supervised learning when small amounts of labeled data are available. Further, we show that Self-Path improves domain adaptation for classification of histology image patches when there is no labeled data available for the target domain. This approach can potentially be employed for other applications in computational pathology, where annotation budget is often limited or large amount of unlabeled image data is available.
Semi-supervised and Unsupervised Methods for Heart Sounds Classification in Restricted Data Environments
Jun 04, 2020


Abstract:Automated heart sounds classification is a much-required diagnostic tool in the view of increasing incidences of heart related diseases worldwide. In this study, we conduct a comprehensive study of heart sounds classification by using various supervised, semi-supervised and unsupervised approaches on the PhysioNet/CinC 2016 Challenge dataset. Supervised approaches, including deep learning and machine learning methods, require large amounts of labelled data to train the models, which are challenging to obtain in most practical scenarios. In view of the need to reduce the labelling burden for clinical practices, where human labelling is both expensive and time-consuming, semi-supervised or even unsupervised approaches in restricted data setting are desirable. A GAN based semi-supervised method is therefore proposed, which allows the usage of unlabelled data samples to boost the learning of data distribution. It achieves a better performance in terms of AUROC over the supervised baseline when limited data samples exist. Furthermore, several unsupervised methods are explored as an alternative approach by considering the given problem as an anomaly detection scenario. In particular, the unsupervised feature extraction using 1D CNN Autoencoder coupled with one-class SVM obtains good performance without any data labelling. The potential of the proposed semi-supervised and unsupervised methods may lead to a workflow tool in the future for the creation of higher quality datasets.
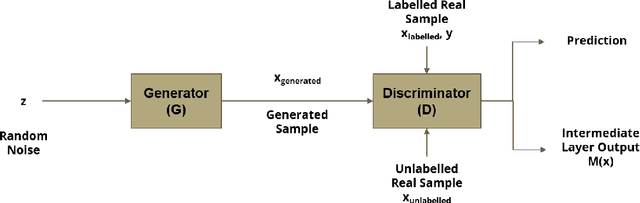
Semi-Supervised Deep Learning for Abnormality Classification in Retinal Images
Dec 19, 2018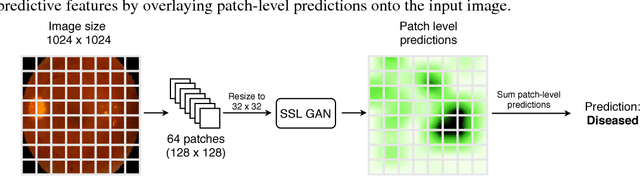


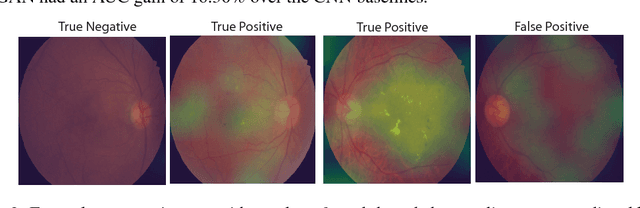
Abstract:Supervised deep learning algorithms have enabled significant performance gains in medical image classification tasks. But these methods rely on large labeled datasets that require resource-intensive expert annotation. Semi-supervised generative adversarial network (GAN) approaches offer a means to learn from limited labeled data alongside larger unlabeled datasets, but have not been applied to discern fine-scale, sparse or localized features that define medical abnormalities. To overcome these limitations, we propose a patch-based semi-supervised learning approach and evaluate performance on classification of diabetic retinopathy from funduscopic images. Our semi-supervised approach achieves high AUC with just 10-20 labeled training images, and outperforms the supervised baselines by upto 15% when less than 30% of the training dataset is labeled. Further, our method implicitly enables interpretation of the SSL predictions. As this approach enables good accuracy, resolution and interpretability with lower annotation burden, it sets the pathway for scalable applications of deep learning in clinical imaging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge