Semi-Supervised Deep Learning for Abnormality Classification in Retinal Images
Paper and Code
Dec 19, 2018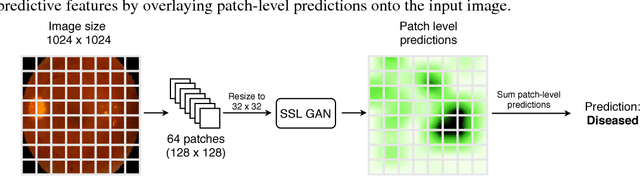


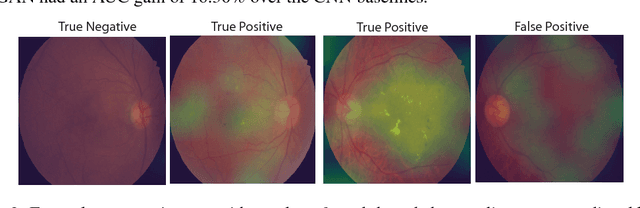
Supervised deep learning algorithms have enabled significant performance gains in medical image classification tasks. But these methods rely on large labeled datasets that require resource-intensive expert annotation. Semi-supervised generative adversarial network (GAN) approaches offer a means to learn from limited labeled data alongside larger unlabeled datasets, but have not been applied to discern fine-scale, sparse or localized features that define medical abnormalities. To overcome these limitations, we propose a patch-based semi-supervised learning approach and evaluate performance on classification of diabetic retinopathy from funduscopic images. Our semi-supervised approach achieves high AUC with just 10-20 labeled training images, and outperforms the supervised baselines by upto 15% when less than 30% of the training dataset is labeled. Further, our method implicitly enables interpretation of the SSL predictions. As this approach enables good accuracy, resolution and interpretability with lower annotation burden, it sets the pathway for scalable applications of deep learning in clinical imaging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge