Tianhao Zhao
A Hierarchical Region-Based Approach for Efficient Multi-Robot Exploration
Mar 17, 2025Abstract:Multi-robot autonomous exploration in an unknown environment is an important application in robotics.Traditional exploration methods only use information around frontier points or viewpoints, ignoring spatial information of unknown areas. Moreover, finding the exact optimal solution for multi-robot task allocation is NP-hard, resulting in significant computational time consumption. To address these issues, we present a hierarchical multi-robot exploration framework using a new modeling method called RegionGraph. The proposed approach makes two main contributions: 1) A new modeling method for unexplored areas that preserves their spatial information across the entire space in a weighted graph called RegionGraph. 2) A hierarchical multi-robot exploration framework that decomposes the global exploration task into smaller subtasks, reducing the frequency of global planning and enabling asynchronous exploration. The proposed method is validated through both simulation and real-world experiments, demonstrating a 20% improvement in efficiency compared to existing methods.
Human Stone Toolmaking Action Grammar (HSTAG): A Challenging Benchmark for Fine-grained Motor Behavior Recognition
Oct 10, 2024Abstract:Action recognition has witnessed the development of a growing number of novel algorithms and datasets in the past decade. However, the majority of public benchmarks were constructed around activities of daily living and annotated at a rather coarse-grained level, which lacks diversity in domain-specific datasets, especially for rarely seen domains. In this paper, we introduced Human Stone Toolmaking Action Grammar (HSTAG), a meticulously annotated video dataset showcasing previously undocumented stone toolmaking behaviors, which can be used for investigating the applications of advanced artificial intelligence techniques in understanding a rapid succession of complex interactions between two hand-held objects. HSTAG consists of 18,739 video clips that record 4.5 hours of experts' activities in stone toolmaking. Its unique features include (i) brief action durations and frequent transitions, mirroring the rapid changes inherent in many motor behaviors; (ii) multiple angles of view and switches among multiple tools, increasing intra-class variability; (iii) unbalanced class distributions and high similarity among different action sequences, adding difficulty in capturing distinct patterns for each action. Several mainstream action recognition models are used to conduct experimental analysis, which showcases the challenges and uniqueness of HSTAG https://nyu.databrary.org/volume/1697.
Improving Bird's Eye View Semantic Segmentation by Task Decomposition
Apr 02, 2024



Abstract:Semantic segmentation in bird's eye view (BEV) plays a crucial role in autonomous driving. Previous methods usually follow an end-to-end pipeline, directly predicting the BEV segmentation map from monocular RGB inputs. However, the challenge arises when the RGB inputs and BEV targets from distinct perspectives, making the direct point-to-point predicting hard to optimize. In this paper, we decompose the original BEV segmentation task into two stages, namely BEV map reconstruction and RGB-BEV feature alignment. In the first stage, we train a BEV autoencoder to reconstruct the BEV segmentation maps given corrupted noisy latent representation, which urges the decoder to learn fundamental knowledge of typical BEV patterns. The second stage involves mapping RGB input images into the BEV latent space of the first stage, directly optimizing the correlations between the two views at the feature level. Our approach simplifies the complexity of combining perception and generation into distinct steps, equipping the model to handle intricate and challenging scenes effectively. Besides, we propose to transform the BEV segmentation map from the Cartesian to the polar coordinate system to establish the column-wise correspondence between RGB images and BEV maps. Moreover, our method requires neither multi-scale features nor camera intrinsic parameters for depth estimation and saves computational overhead. Extensive experiments on nuScenes and Argoverse show the effectiveness and efficiency of our method. Code is available at https://github.com/happytianhao/TaDe.
MixSiam: A Mixture-based Approach to Self-supervised Representation Learning
Nov 04, 2021



Abstract:Recently contrastive learning has shown significant progress in learning visual representations from unlabeled data. The core idea is training the backbone to be invariant to different augmentations of an instance. While most methods only maximize the feature similarity between two augmented data, we further generate more challenging training samples and force the model to keep predicting discriminative representation on these hard samples. In this paper, we propose MixSiam, a mixture-based approach upon the traditional siamese network. On the one hand, we input two augmented images of an instance to the backbone and obtain the discriminative representation by performing an element-wise maximum of two features. On the other hand, we take the mixture of these augmented images as input, and expect the model prediction to be close to the discriminative representation. In this way, the model could access more variant data samples of an instance and keep predicting invariant discriminative representations for them. Thus the learned model is more robust compared to previous contrastive learning methods. Extensive experiments on large-scale datasets show that MixSiam steadily improves the baseline and achieves competitive results with state-of-the-art methods. Our code will be released soon.
Learning from Thresholds: Fully Automated Classification of Tumor Infiltrating Lymphocytes for Multiple Cancer Types
Jul 09, 2019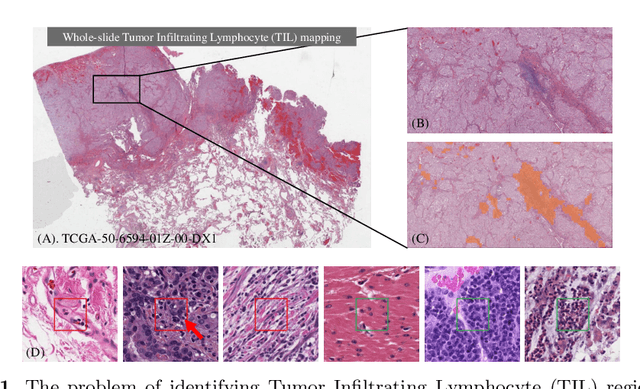

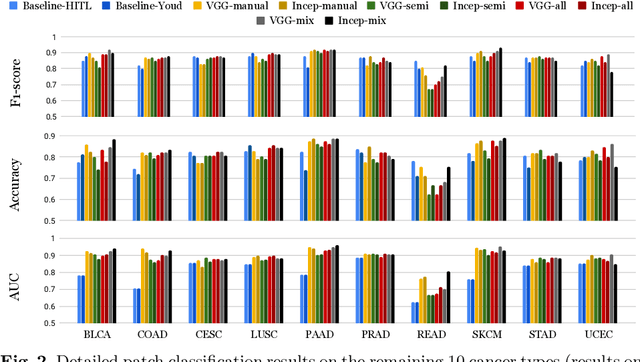
Abstract:Deep learning classifiers for characterization of whole slide tissue morphology require large volumes of annotated data to learn variations across different tissue and cancer types. As is well known, manual generation of digital pathology training data is time consuming and expensive. In this paper, we propose a semi-automated method for annotating a group of similar instances at once, instead of collecting only per-instance manual annotations. This allows for a much larger training set, that reflects visual variability across multiple cancer types and thus training of a single network which can be automatically applied to each cancer type without human adjustment. We apply our method to the important task of classifying Tumor Infiltrating Lymphocytes (TILs) in H&E images. Prior approaches were trained for individual cancer types, with smaller training sets and human-in-the-loop threshold adjustment. We utilize these thresholded results as large scale "semi-automatic" annotations. Combined with existing manual annotations, our trained deep networks are able to automatically produce better TIL prediction results in 12 cancer types, compared to the human-in-the-loop approach.
Utilizing Automated Breast Cancer Detection to Identify Spatial Distributions of Tumor Infiltrating Lymphocytes in Invasive Breast Cancer
May 29, 2019
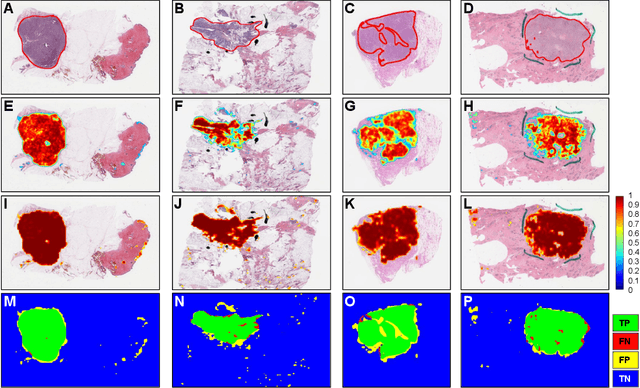
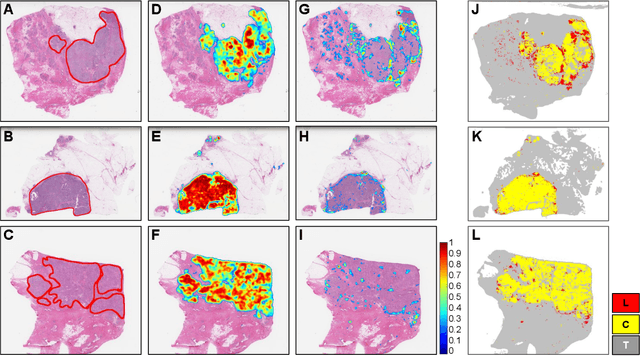
Abstract:Quantitative assessment of Tumor-TIL spatial relationships is increasingly important in both basic science and clinical aspects of breast cancer research. We have developed and evaluated convolutional neural network (CNN) analysis pipelines to generate combined maps of cancer regions and tumor infiltrating lymphocytes (TILs) in routine diagnostic breast cancer whole slide tissue images (WSIs). We produce interactive whole slide maps that provide 1) insight about the structural patterns and spatial distribution of lymphocytic infiltrates and 2) facilitate improved quantification of TILs. We evaluated both tumor and TIL analyses using three CNN networks - Resnet-34, VGG16 and Inception v4, and demonstrated that the results compared favorably to those obtained by what believe are the best published methods. We have produced open-source tools and generated a public dataset consisting of tumor/TIL maps for 1,015 TCGA breast cancer images. We also present a customized web-based interface that enables easy visualization and interactive exploration of high-resolution combined Tumor-TIL maps for 1,015TCGA invasive breast cancer cases that can be downloaded for further downstream analyses.
Methods for Segmentation and Classification of Digital Microscopy Tissue Images
Oct 31, 2018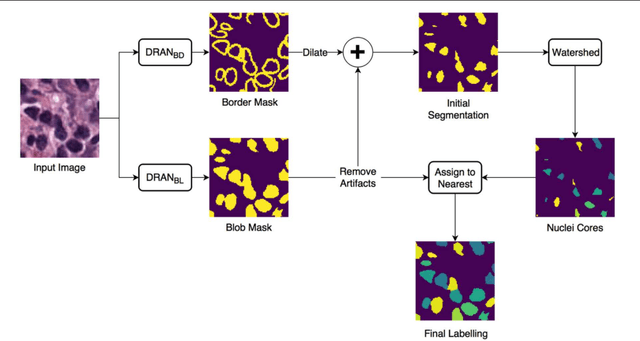

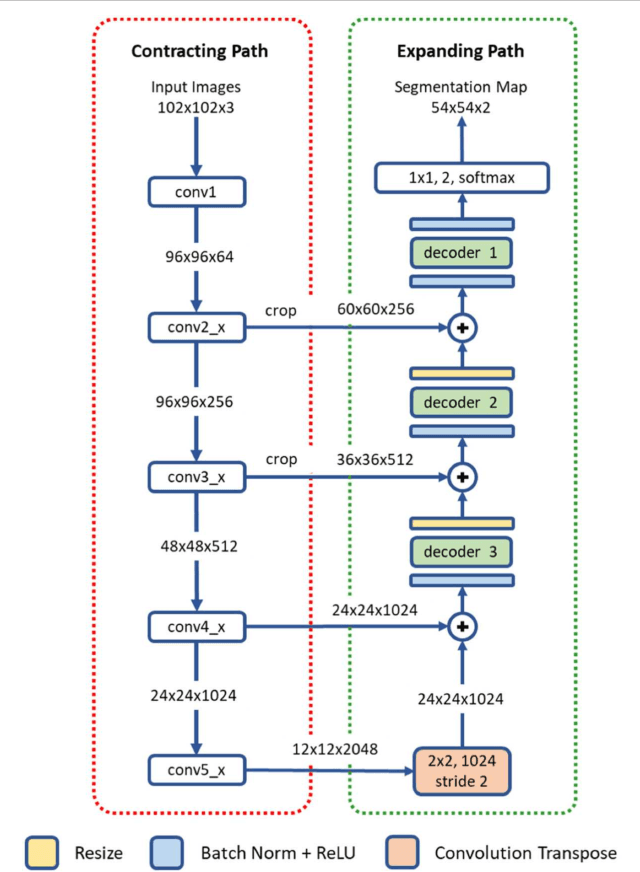
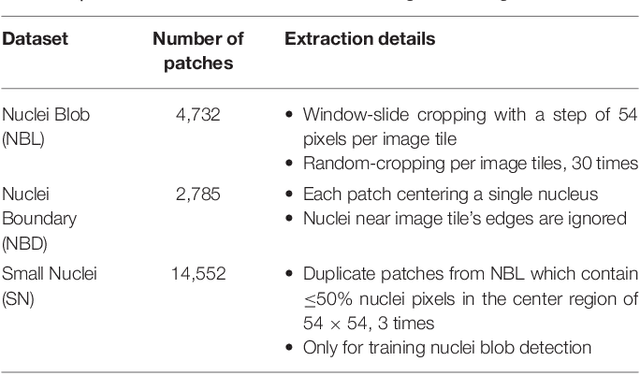
Abstract:High-resolution microscopy images of tissue specimens provide detailed information about the morphology of normal and diseased tissue. Image analysis of tissue morphology can help cancer researchers develop a better understanding of cancer biology. Segmentation of nuclei and classification of tissue images are two common tasks in tissue image analysis. Development of accurate and efficient algorithms for these tasks is a challenging problem because of the complexity of tissue morphology and tumor heterogeneity. In this paper we present two computer algorithms; one designed for segmentation of nuclei and the other for classification of whole slide tissue images. The segmentation algorithm implements a multiscale deep residual aggregation network to accurately segment nuclear material and then separate clumped nuclei into individual nuclei. The classification algorithm initially carries out patch-level classification via a deep learning method, then patch-level statistical and morphological features are used as input to a random forest regression model for whole slide image classification. The segmentation and classification algorithms were evaluated in the MICCAI 2017 Digital Pathology challenge. The segmentation algorithm achieved an accuracy score of 0.78. The classification algorithm achieved an accuracy score of 0.81.
3D Facial Expression Reconstruction using Cascaded Regression
Aug 17, 2018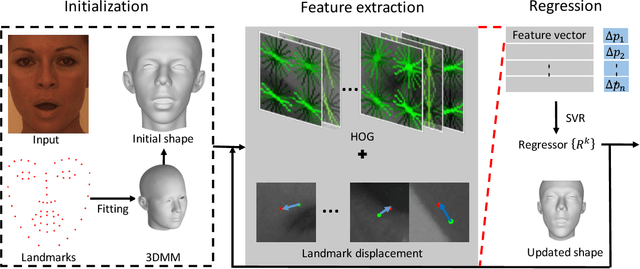
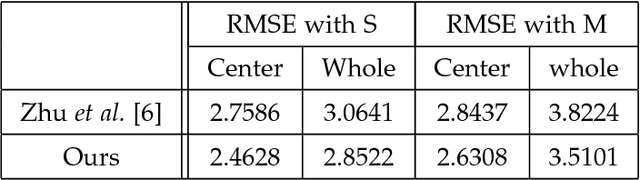
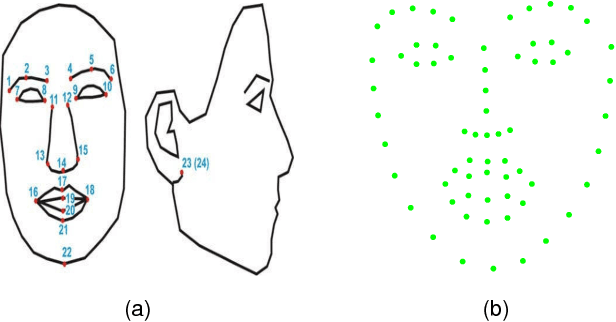
Abstract:This paper proposes a novel model fitting algorithm for 3D facial expression reconstruction from a single image. Face expression reconstruction from a single image is a challenging task in computer vision. Most state-of-the-art methods fit the input image to a 3D Morphable Model (3DMM). These methods need to solve a stochastic problem and cannot deal with expression and pose variations. To solve this problem, we adopt a 3D face expression model and use a combined feature which is robust to scale, rotation and different lighting conditions. The proposed method applies a cascaded regression framework to estimate parameters for the 3DMM. 2D landmarks are detected and used to initialize the 3D shape and mapping matrices. In each iteration, residues between the current 3DMM parameters and the ground truth are estimated and then used to update the 3D shapes. The mapping matrices are also calculated based on the updated shapes and 2D landmarks. HOG features of the local patches and displacements between 3D landmark projections and 2D landmarks are exploited. Compared with existing methods, the proposed method is robust to expression and pose changes and can reconstruct higher fidelity 3D face shape.
Sparse Autoencoder for Unsupervised Nucleus Detection and Representation in Histopathology Images
Apr 10, 2017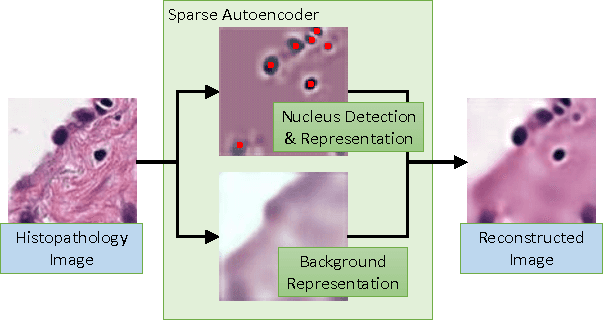
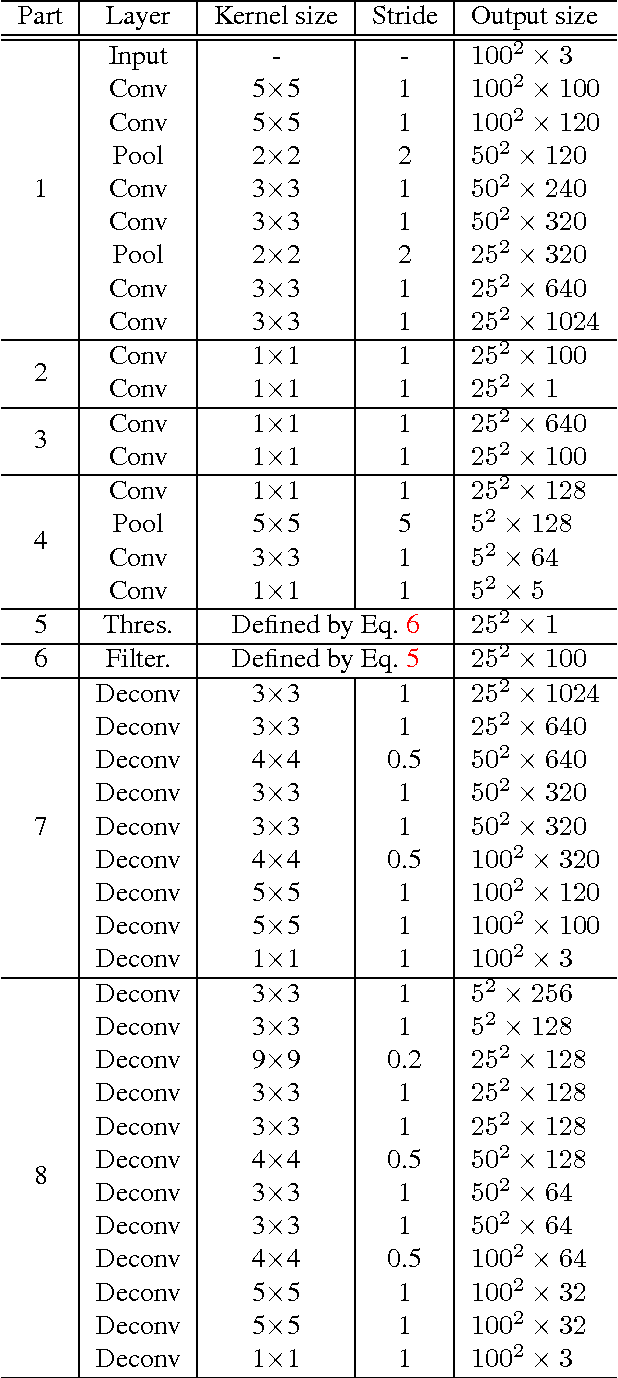
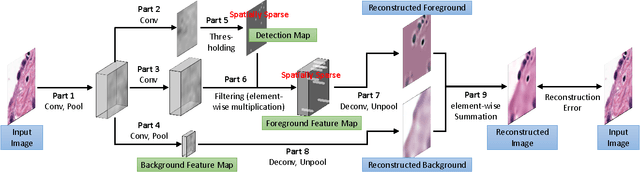
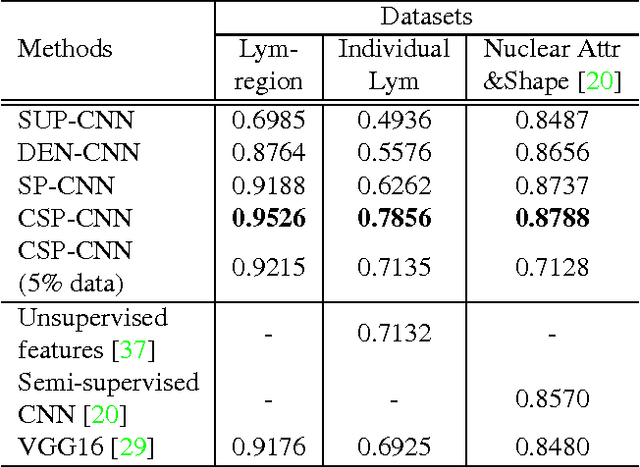
Abstract:Histopathology images are crucial to the study of complex diseases such as cancer. The histologic characteristics of nuclei play a key role in disease diagnosis, prognosis and analysis. In this work, we propose a sparse Convolutional Autoencoder (CAE) for fully unsupervised, simultaneous nucleus detection and feature extraction in histopathology tissue images. Our CAE detects and encodes nuclei in image patches in tissue images into sparse feature maps that encode both the location and appearance of nuclei. Our CAE is the first unsupervised detection network for computer vision applications. The pretrained nucleus detection and feature extraction modules in our CAE can be fine-tuned for supervised learning in an end-to-end fashion. We evaluate our method on four datasets and reduce the errors of state-of-the-art methods up to 42%. We are able to achieve comparable performance with only 5% of the fully-supervised annotation cost.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge