Rajarsi Gupta
$\infty$-Brush: Controllable Large Image Synthesis with Diffusion Models in Infinite Dimensions
Jul 20, 2024



Abstract:Synthesizing high-resolution images from intricate, domain-specific information remains a significant challenge in generative modeling, particularly for applications in large-image domains such as digital histopathology and remote sensing. Existing methods face critical limitations: conditional diffusion models in pixel or latent space cannot exceed the resolution on which they were trained without losing fidelity, and computational demands increase significantly for larger image sizes. Patch-based methods offer computational efficiency but fail to capture long-range spatial relationships due to their overreliance on local information. In this paper, we introduce a novel conditional diffusion model in infinite dimensions, $\infty$-Brush for controllable large image synthesis. We propose a cross-attention neural operator to enable conditioning in function space. Our model overcomes the constraints of traditional finite-dimensional diffusion models and patch-based methods, offering scalability and superior capability in preserving global image structures while maintaining fine details. To our best knowledge, $\infty$-Brush is the first conditional diffusion model in function space, that can controllably synthesize images at arbitrary resolutions of up to $4096\times4096$ pixels. The code is available at https://github.com/cvlab-stonybrook/infinity-brush.
Decoding the visual attention of pathologists to reveal their level of expertise
Mar 25, 2024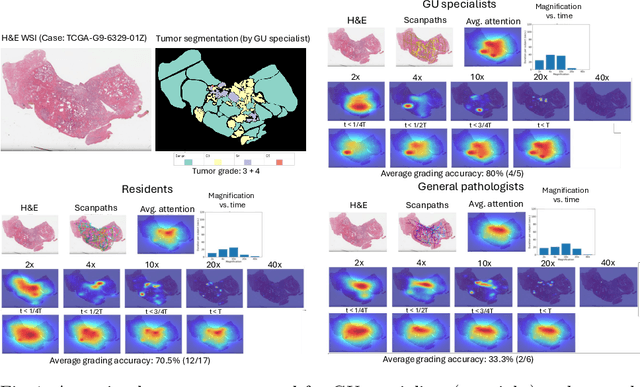
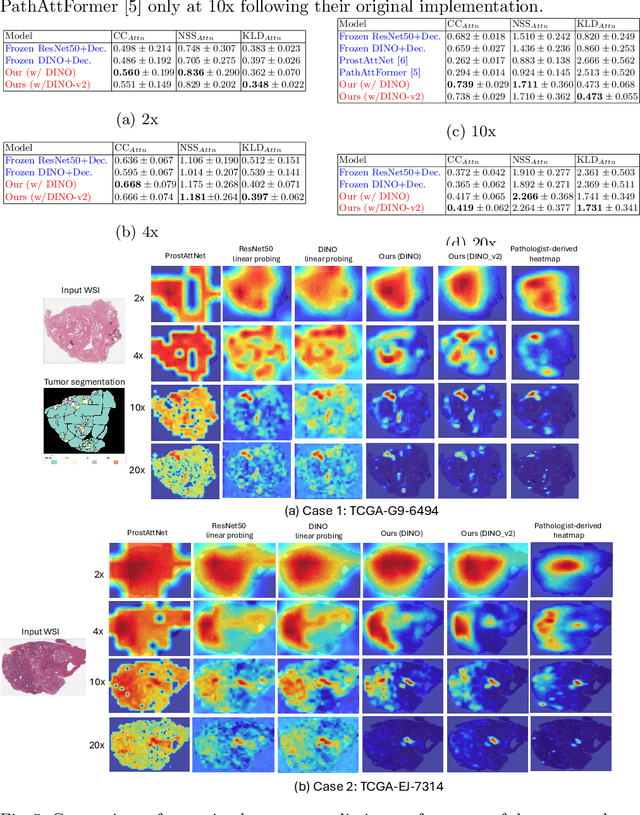
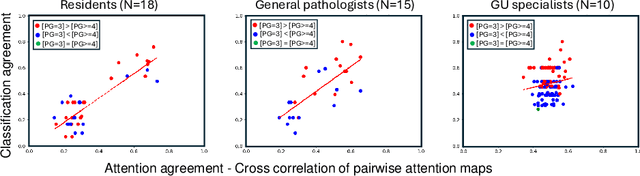
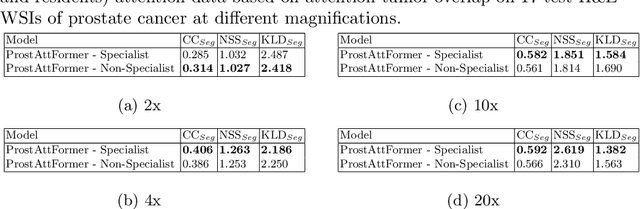
Abstract:We present a method for classifying the expertise of a pathologist based on how they allocated their attention during a cancer reading. We engage this decoding task by developing a novel method for predicting the attention of pathologists as they read whole-slide Images (WSIs) of prostate and make cancer grade classifications. Our ground truth measure of a pathologists' attention is the x, y and z (magnification) movement of their viewport as they navigated through WSIs during readings, and to date we have the attention behavior of 43 pathologists reading 123 WSIs. These data revealed that specialists have higher agreement in both their attention and cancer grades compared to general pathologists and residents, suggesting that sufficient information may exist in their attention behavior to classify their expertise level. To attempt this, we trained a transformer-based model to predict the visual attention heatmaps of resident, general, and specialist (GU) pathologists during Gleason grading. Based solely on a pathologist's attention during a reading, our model was able to predict their level of expertise with 75.3%, 56.1%, and 77.2% accuracy, respectively, better than chance and baseline models. Our model therefore enables a pathologist's expertise level to be easily and objectively evaluated, important for pathology training and competency assessment. Tools developed from our model could also be used to help pathology trainees learn how to read WSIs like an expert.
Open and reusable deep learning for pathology with WSInfer and QuPath
Sep 08, 2023Abstract:The field of digital pathology has seen a proliferation of deep learning models in recent years. Despite substantial progress, it remains rare for other researchers and pathologists to be able to access models published in the literature and apply them to their own images. This is due to difficulties in both sharing and running models. To address these concerns, we introduce WSInfer: a new, open-source software ecosystem designed to make deep learning for pathology more streamlined and accessible. WSInfer comprises three main elements: 1) a Python package and command line tool to efficiently apply patch-based deep learning inference to whole slide images; 2) a QuPath extension that provides an alternative inference engine through user-friendly and interactive software, and 3) a model zoo, which enables pathology models and metadata to be easily shared in a standardized form. Together, these contributions aim to encourage wider reuse, exploration, and interrogation of deep learning models for research purposes, by putting them into the hands of pathologists and eliminating a need for coding experience when accessed through QuPath. The WSInfer source code is hosted on GitHub and documentation is available at https://wsinfer.readthedocs.io.
Halcyon -- A Pathology Imaging and Feature analysis and Management System
Apr 07, 2023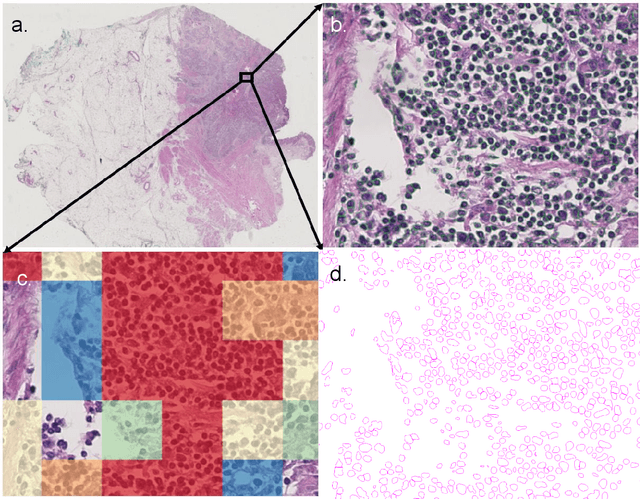
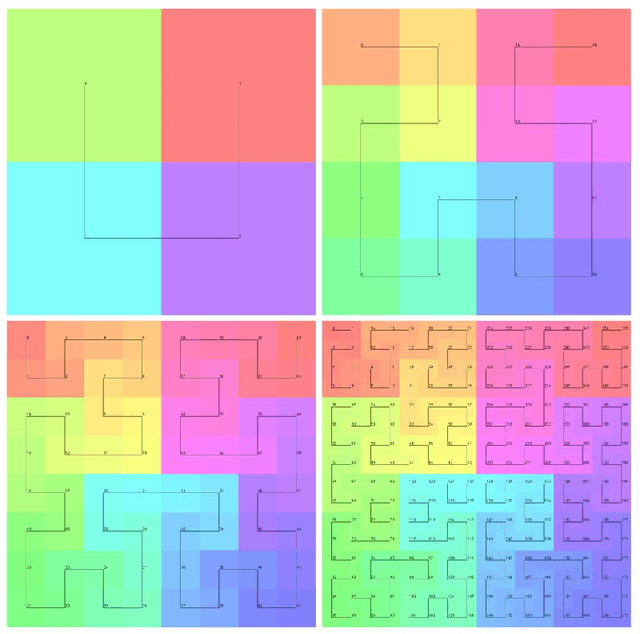
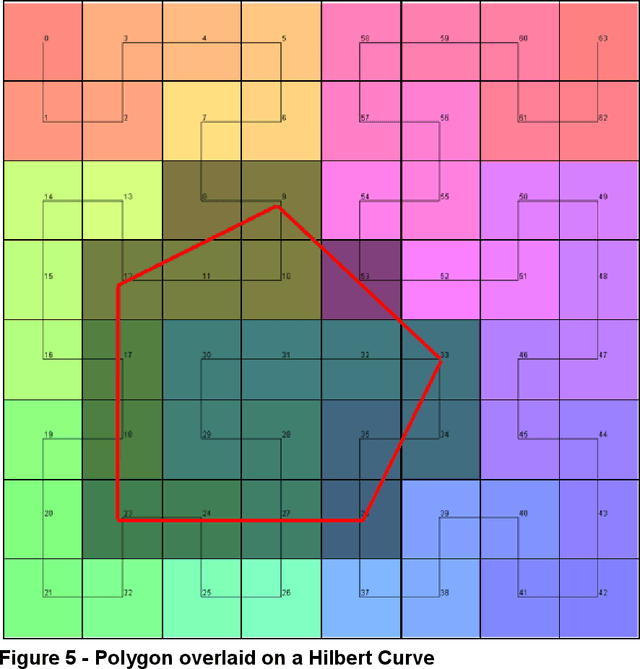
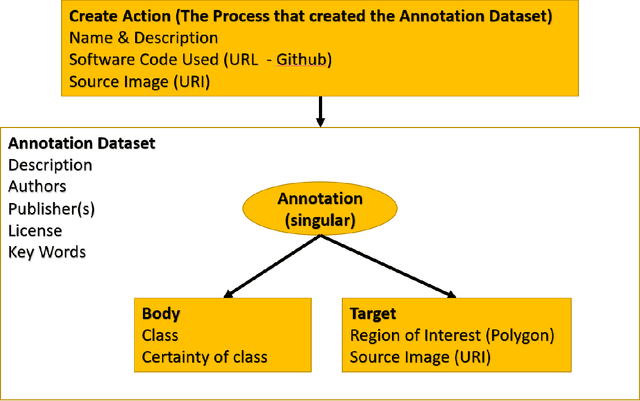
Abstract:Halcyon is a new pathology imaging analysis and feature management system based on W3C linked-data open standards and is designed to scale to support the needs for the voluminous production of features from deep-learning feature pipelines. Halcyon can support multiple users with a web-based UX with access to all user data over a standards-based web API allowing for integration with other processes and software systems. Identity management and data security is also provided.
Topology-Guided Multi-Class Cell Context Generation for Digital Pathology
Apr 05, 2023



Abstract:In digital pathology, the spatial context of cells is important for cell classification, cancer diagnosis and prognosis. To model such complex cell context, however, is challenging. Cells form different mixtures, lineages, clusters and holes. To model such structural patterns in a learnable fashion, we introduce several mathematical tools from spatial statistics and topological data analysis. We incorporate such structural descriptors into a deep generative model as both conditional inputs and a differentiable loss. This way, we are able to generate high quality multi-class cell layouts for the first time. We show that the topology-rich cell layouts can be used for data augmentation and improve the performance of downstream tasks such as cell classification.
ViT-DAE: Transformer-driven Diffusion Autoencoder for Histopathology Image Analysis
Apr 03, 2023Abstract:Generative AI has received substantial attention in recent years due to its ability to synthesize data that closely resembles the original data source. While Generative Adversarial Networks (GANs) have provided innovative approaches for histopathological image analysis, they suffer from limitations such as mode collapse and overfitting in discriminator. Recently, Denoising Diffusion models have demonstrated promising results in computer vision. These models exhibit superior stability during training, better distribution coverage, and produce high-quality diverse images. Additionally, they display a high degree of resilience to noise and perturbations, making them well-suited for use in digital pathology, where images commonly contain artifacts and exhibit significant variations in staining. In this paper, we present a novel approach, namely ViT-DAE, which integrates vision transformers (ViT) and diffusion autoencoders for high-quality histopathology image synthesis. This marks the first time that ViT has been introduced to diffusion autoencoders in computational pathology, allowing the model to better capture the complex and intricate details of histopathology images. We demonstrate the effectiveness of ViT-DAE on three publicly available datasets. Our approach outperforms recent GAN-based and vanilla DAE methods in generating realistic images.
Gigapixel Whole-Slide Images Classification using Locally Supervised Learning
Jul 17, 2022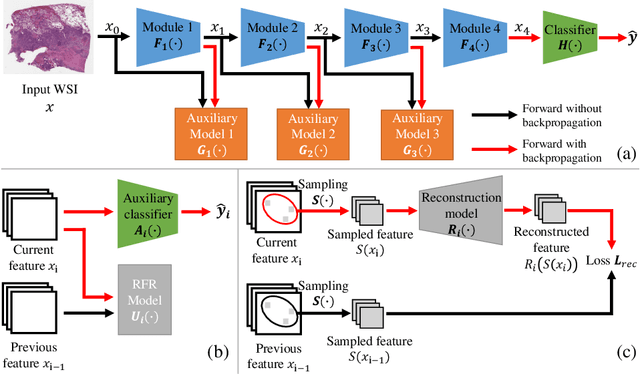
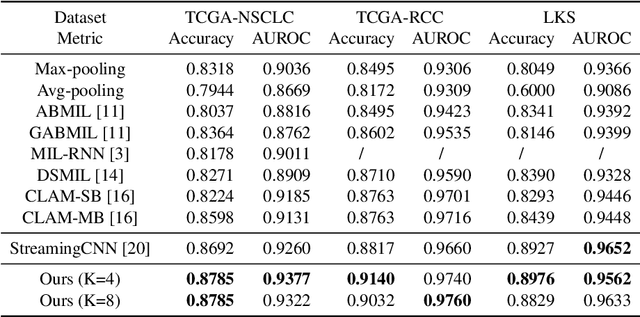

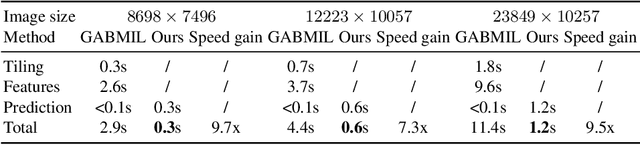
Abstract:Histopathology whole slide images (WSIs) play a very important role in clinical studies and serve as the gold standard for many cancer diagnoses. However, generating automatic tools for processing WSIs is challenging due to their enormous sizes. Currently, to deal with this issue, conventional methods rely on a multiple instance learning (MIL) strategy to process a WSI at patch level. Although effective, such methods are computationally expensive, because tiling a WSI into patches takes time and does not explore the spatial relations between these tiles. To tackle these limitations, we propose a locally supervised learning framework which processes the entire slide by exploring the entire local and global information that it contains. This framework divides a pre-trained network into several modules and optimizes each module locally using an auxiliary model. We also introduce a random feature reconstruction unit (RFR) to preserve distinguishing features during training and improve the performance of our method by 1% to 3%. Extensive experiments on three publicly available WSI datasets: TCGA-NSCLC, TCGA-RCC and LKS, highlight the superiority of our method on different classification tasks. Our method outperforms the state-of-the-art MIL methods by 2% to 5% in accuracy, while being 7 to 10 times faster. Additionally, when dividing it into eight modules, our method requires as little as 20% of the total gpu memory required by end-to-end training. Our code is available at https://github.com/cvlab-stonybrook/local_learning_wsi.
AI and Pathology: Steering Treatment and Predicting Outcomes
Jun 15, 2022
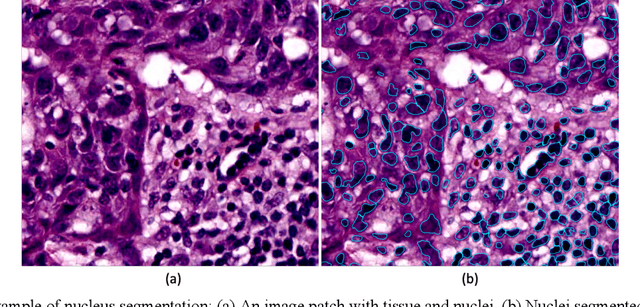
Abstract:The combination of data analysis methods, increasing computing capacity, and improved sensors enable quantitative granular, multi-scale, cell-based analyses. We describe the rich set of application challenges related to tissue interpretation and survey AI methods currently used to address these challenges. We focus on a particular class of targeted human tissue analysis - histopathology - aimed at quantitative characterization of disease state, patient outcome prediction and treatment steering.
Evaluating histopathology transfer learning with ChampKit
Jun 14, 2022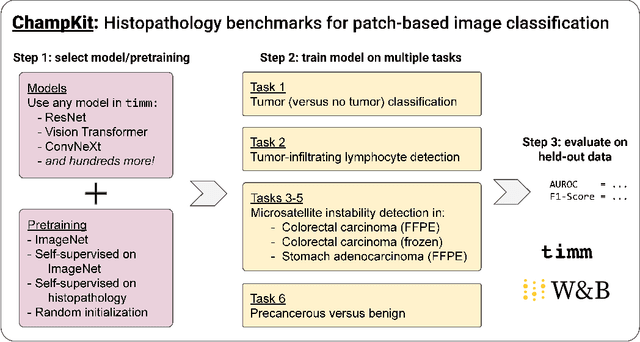
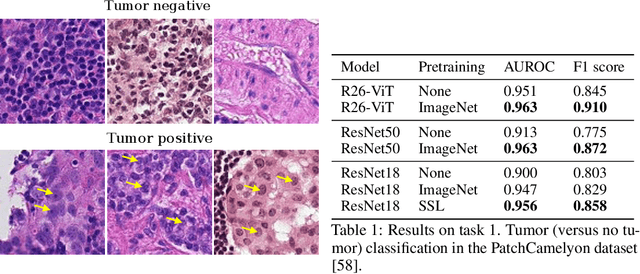
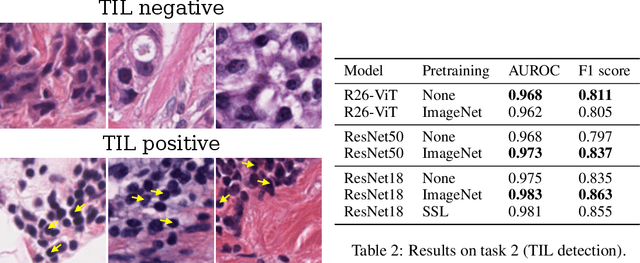
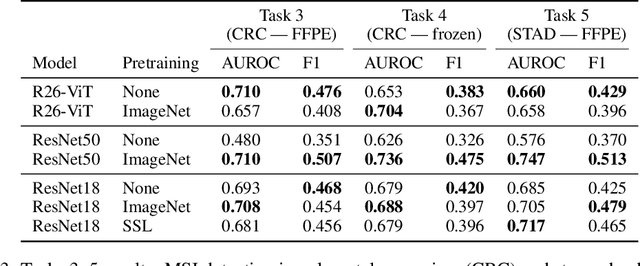
Abstract:Histopathology remains the gold standard for diagnosis of various cancers. Recent advances in computer vision, specifically deep learning, have facilitated the analysis of histopathology images for various tasks, including immune cell detection and microsatellite instability classification. The state-of-the-art for each task often employs base architectures that have been pretrained for image classification on ImageNet. The standard approach to develop classifiers in histopathology tends to focus narrowly on optimizing models for a single task, not considering the aspects of modeling innovations that improve generalization across tasks. Here we present ChampKit (Comprehensive Histopathology Assessment of Model Predictions toolKit): an extensible, fully reproducible benchmarking toolkit that consists of a broad collection of patch-level image classification tasks across different cancers. ChampKit enables a way to systematically document the performance impact of proposed improvements in models and methodology. ChampKit source code and data are freely accessible at https://github.com/kaczmarj/champkit .
Federated Learning for the Classification of Tumor Infiltrating Lymphocytes
Apr 01, 2022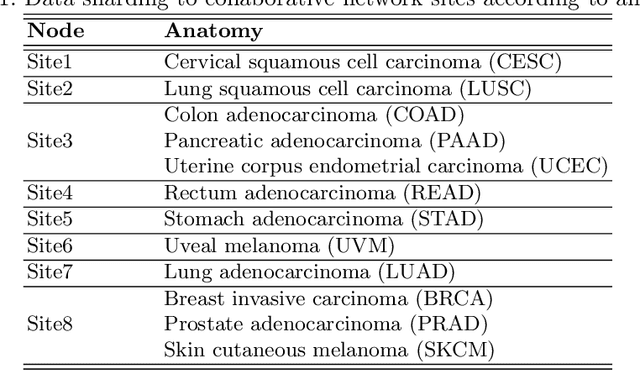
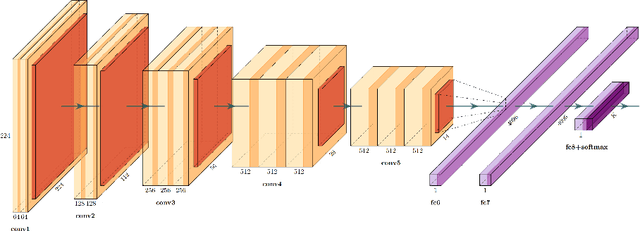
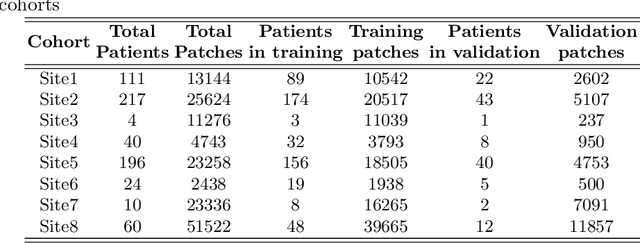

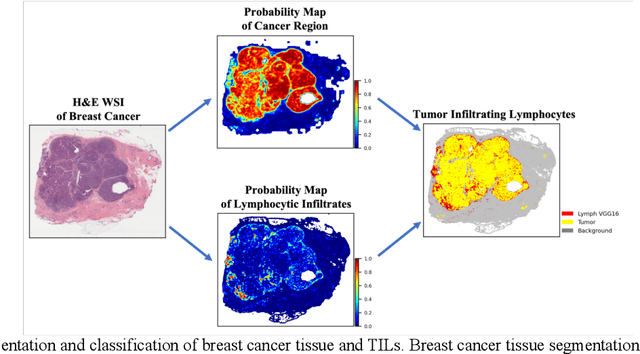
Abstract:We evaluate the performance of federated learning (FL) in developing deep learning models for analysis of digitized tissue sections. A classification application was considered as the example use case, on quantifiying the distribution of tumor infiltrating lymphocytes within whole slide images (WSIs). A deep learning classification model was trained using 50*50 square micron patches extracted from the WSIs. We simulated a FL environment in which a dataset, generated from WSIs of cancer from numerous anatomical sites available by The Cancer Genome Atlas repository, is partitioned in 8 different nodes. Our results show that the model trained with the federated training approach achieves similar performance, both quantitatively and qualitatively, to that of a model trained with all the training data pooled at a centralized location. Our study shows that FL has tremendous potential for enabling development of more robust and accurate models for histopathology image analysis without having to collect large and diverse training data at a single location.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge