Erich Bremer
Open and reusable deep learning for pathology with WSInfer and QuPath
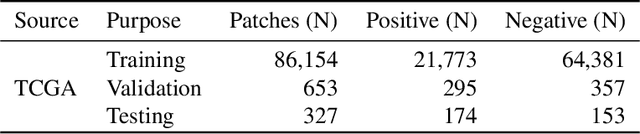
Sep 08, 2023Abstract:The field of digital pathology has seen a proliferation of deep learning models in recent years. Despite substantial progress, it remains rare for other researchers and pathologists to be able to access models published in the literature and apply them to their own images. This is due to difficulties in both sharing and running models. To address these concerns, we introduce WSInfer: a new, open-source software ecosystem designed to make deep learning for pathology more streamlined and accessible. WSInfer comprises three main elements: 1) a Python package and command line tool to efficiently apply patch-based deep learning inference to whole slide images; 2) a QuPath extension that provides an alternative inference engine through user-friendly and interactive software, and 3) a model zoo, which enables pathology models and metadata to be easily shared in a standardized form. Together, these contributions aim to encourage wider reuse, exploration, and interrogation of deep learning models for research purposes, by putting them into the hands of pathologists and eliminating a need for coding experience when accessed through QuPath. The WSInfer source code is hosted on GitHub and documentation is available at https://wsinfer.readthedocs.io.
Halcyon -- A Pathology Imaging and Feature analysis and Management System
Apr 07, 2023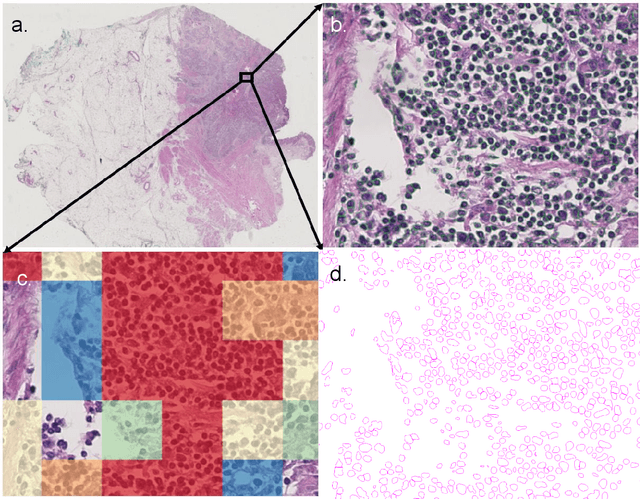
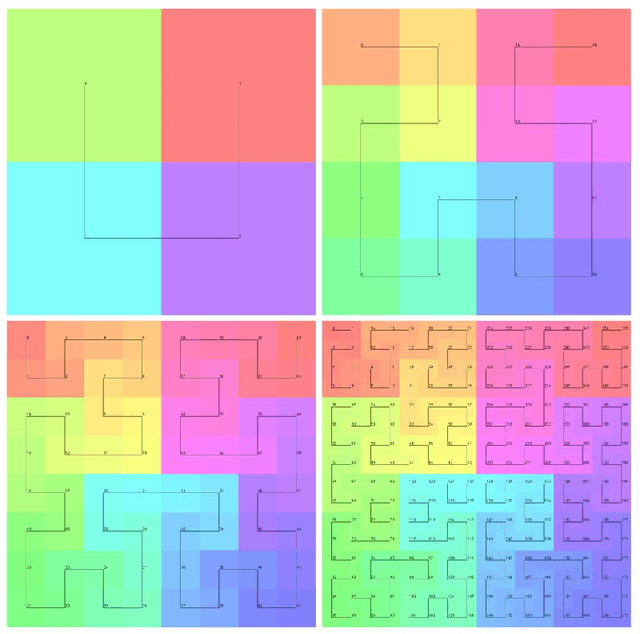
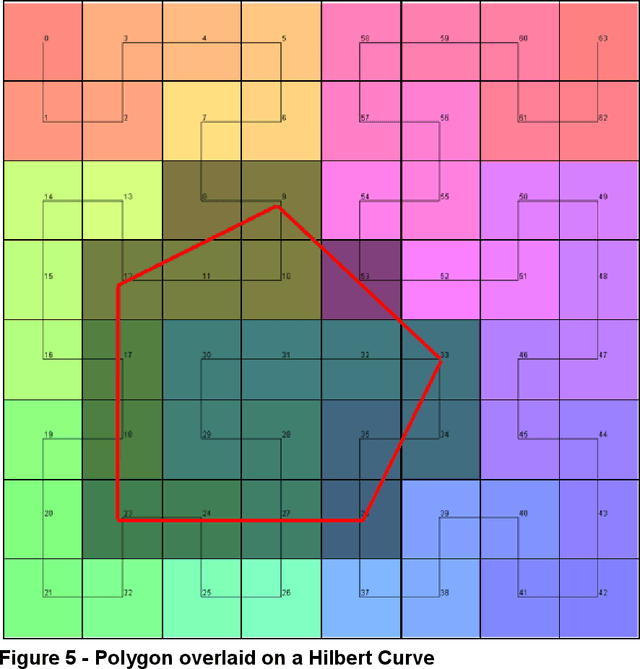
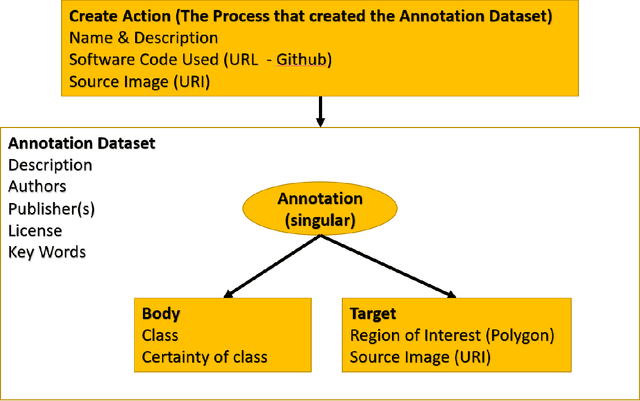
Abstract:Halcyon is a new pathology imaging analysis and feature management system based on W3C linked-data open standards and is designed to scale to support the needs for the voluminous production of features from deep-learning feature pipelines. Halcyon can support multiple users with a web-based UX with access to all user data over a standards-based web API allowing for integration with other processes and software systems. Identity management and data security is also provided.
ImageBox3: No-Server Tile Serving to Traverse Whole Slide Images on the Web
Jul 06, 2022Abstract:Whole slide imaging (WSI) has become the primary modality for digital pathology data. However, due to the size and high-resolution nature of these images, they are generally only accessed in smaller sections or tiles via specialized platforms, most of which require extensive setup and/or costly infrastructure. These platforms typically also need a copy of the images to be locally available to them, potentially causing issues with data governance and provenance. To address these concerns, we developed ImageBox3, an in-browser tiling mechanism to enable zero-footprint traversal of remote WSI data. All computation is performed client-side without compromising user governance, operating public and private images alike as long as the storage service supports HTTP range requests (standard in Cloud storage and most web servers). ImageBox3 thus removes significant hurdles to WSI operation and effective collaboration, allowing for the sort of democratized analytical tools needed to establish participative, FAIR digital pathology data commons. Availability: code - https://github.com/episphere/imagebox3; fig1 (live) - https://episphere.github.io/imagebox3/demo/scriptTag ; fig2 (live) - https://episphere.github.io/imagebox3/demo/serviceWorker ; fig 3 (live) - https://observablehq.com/@prafulb/imagebox3-in-observable .
Federated Learning for the Classification of Tumor Infiltrating Lymphocytes
Apr 01, 2022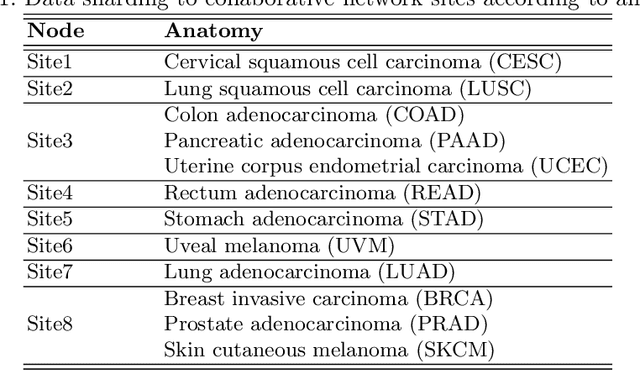
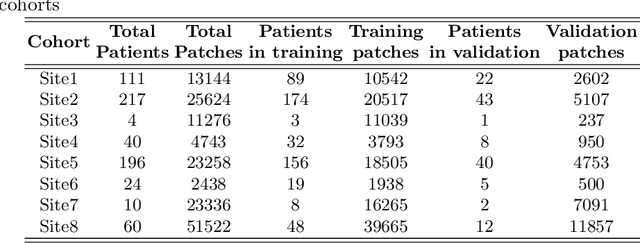

Abstract:We evaluate the performance of federated learning (FL) in developing deep learning models for analysis of digitized tissue sections. A classification application was considered as the example use case, on quantifiying the distribution of tumor infiltrating lymphocytes within whole slide images (WSIs). A deep learning classification model was trained using 50*50 square micron patches extracted from the WSIs. We simulated a FL environment in which a dataset, generated from WSIs of cancer from numerous anatomical sites available by The Cancer Genome Atlas repository, is partitioned in 8 different nodes. Our results show that the model trained with the federated training approach achieves similar performance, both quantitatively and qualitatively, to that of a model trained with all the training data pooled at a centralized location. Our study shows that FL has tremendous potential for enabling development of more robust and accurate models for histopathology image analysis without having to collect large and diverse training data at a single location.
Utilizing Automated Breast Cancer Detection to Identify Spatial Distributions of Tumor Infiltrating Lymphocytes in Invasive Breast Cancer
May 29, 2019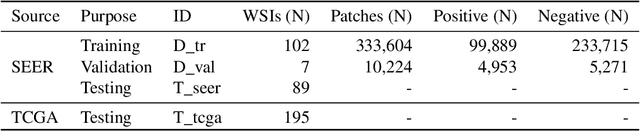
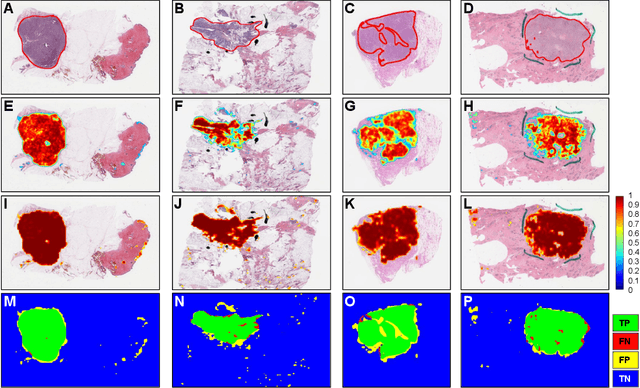
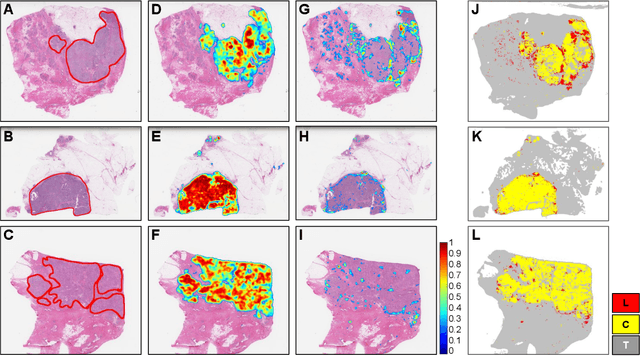
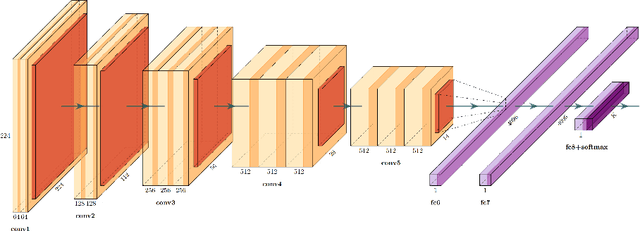
Abstract:Quantitative assessment of Tumor-TIL spatial relationships is increasingly important in both basic science and clinical aspects of breast cancer research. We have developed and evaluated convolutional neural network (CNN) analysis pipelines to generate combined maps of cancer regions and tumor infiltrating lymphocytes (TILs) in routine diagnostic breast cancer whole slide tissue images (WSIs). We produce interactive whole slide maps that provide 1) insight about the structural patterns and spatial distribution of lymphocytic infiltrates and 2) facilitate improved quantification of TILs. We evaluated both tumor and TIL analyses using three CNN networks - Resnet-34, VGG16 and Inception v4, and demonstrated that the results compared favorably to those obtained by what believe are the best published methods. We have produced open-source tools and generated a public dataset consisting of tumor/TIL maps for 1,015 TCGA breast cancer images. We also present a customized web-based interface that enables easy visualization and interactive exploration of high-resolution combined Tumor-TIL maps for 1,015TCGA invasive breast cancer cases that can be downloaded for further downstream analyses.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge