Praphulla MS Bhawsar
TMA-Grid: An open-source, zero-footprint web application for FAIR Tissue MicroArray De-arraying
Jul 30, 2024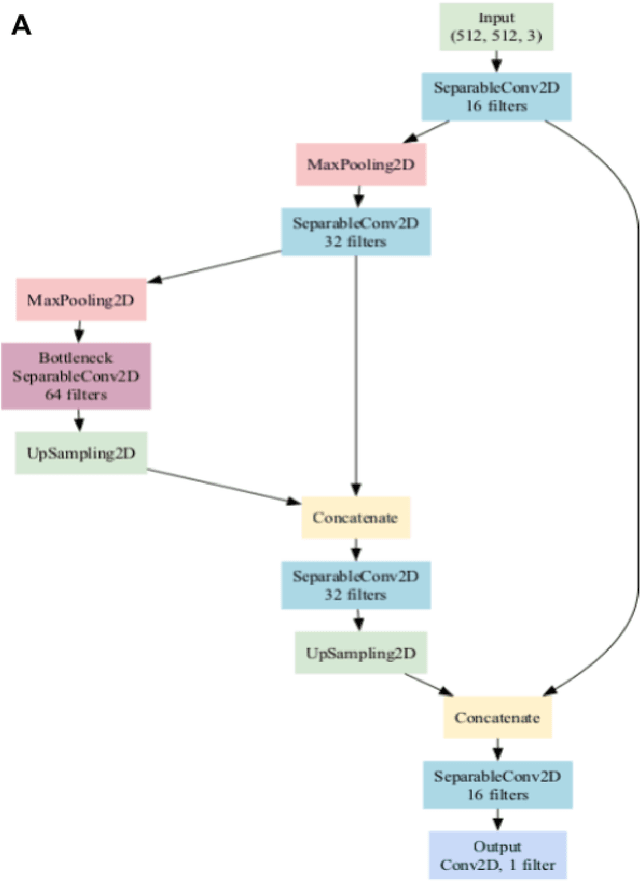

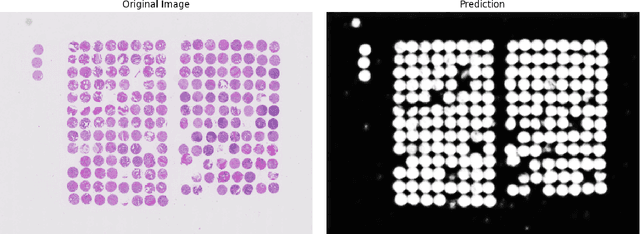
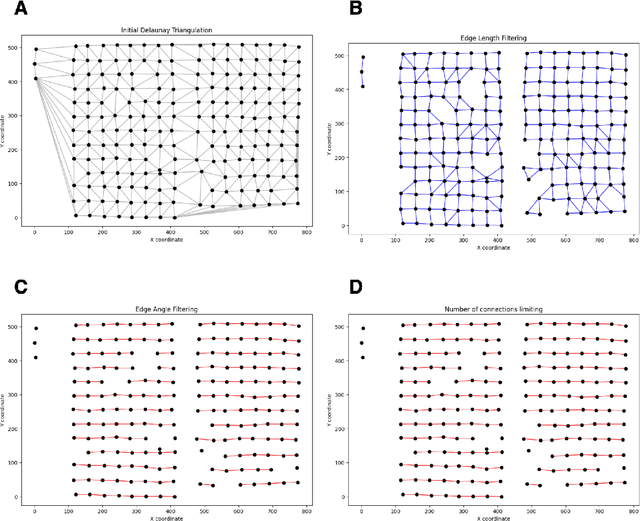
Abstract:Background: Tissue Microarrays (TMAs) significantly increase analytical efficiency in histopathology and large-scale epidemiologic studies by allowing multiple tissue cores to be scanned on a single slide. The individual cores can be digitally extracted and then linked to metadata for analysis in a process known as de-arraying. However, TMAs often contain core misalignments and artifacts due to assembly errors, which can adversely affect the reliability of the extracted cores during the de-arraying process. Moreover, conventional approaches for TMA de-arraying rely on desktop solutions.Therefore, a robust yet flexible de-arraying method is crucial to account for these inaccuracies and ensure effective downstream analyses. Results: We developed TMA-Grid, an in-browser, zero-footprint, interactive web application for TMA de-arraying. This web application integrates a convolutional neural network for precise tissue segmentation and a grid estimation algorithm to match each identified core to its expected location. The application emphasizes interactivity, allowing users to easily adjust segmentation and gridding results. Operating entirely in the web-browser, TMA-Grid eliminates the need for downloads or installations and ensures data privacy. Adhering to FAIR principles (Findable, Accessible, Interoperable, and Reusable), the application and its components are designed for seamless integration into TMA research workflows. Conclusions: TMA-Grid provides a robust, user-friendly solution for TMA dearraying on the web. As an open, freely accessible platform, it lays the foundation for collaborative analyses of TMAs and similar histopathology imaging data. Availability: Web application: https://episphere.github.io/tma-grid Code: https://github.com/episphere/tma-grid Tutorial: https://youtu.be/miajqyw4BVk
ImageBox3: No-Server Tile Serving to Traverse Whole Slide Images on the Web
Jul 06, 2022Abstract:Whole slide imaging (WSI) has become the primary modality for digital pathology data. However, due to the size and high-resolution nature of these images, they are generally only accessed in smaller sections or tiles via specialized platforms, most of which require extensive setup and/or costly infrastructure. These platforms typically also need a copy of the images to be locally available to them, potentially causing issues with data governance and provenance. To address these concerns, we developed ImageBox3, an in-browser tiling mechanism to enable zero-footprint traversal of remote WSI data. All computation is performed client-side without compromising user governance, operating public and private images alike as long as the storage service supports HTTP range requests (standard in Cloud storage and most web servers). ImageBox3 thus removes significant hurdles to WSI operation and effective collaboration, allowing for the sort of democratized analytical tools needed to establish participative, FAIR digital pathology data commons. Availability: code - https://github.com/episphere/imagebox3; fig1 (live) - https://episphere.github.io/imagebox3/demo/scriptTag ; fig2 (live) - https://episphere.github.io/imagebox3/demo/serviceWorker ; fig 3 (live) - https://observablehq.com/@prafulb/imagebox3-in-observable .
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge