Danielle Fassler
Utilizing Automated Breast Cancer Detection to Identify Spatial Distributions of Tumor Infiltrating Lymphocytes in Invasive Breast Cancer
May 29, 2019
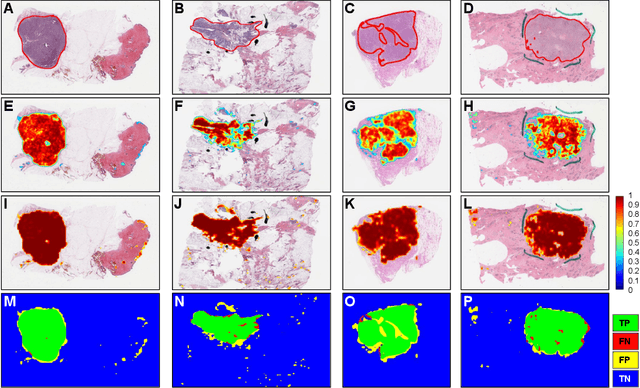
Abstract:Quantitative assessment of Tumor-TIL spatial relationships is increasingly important in both basic science and clinical aspects of breast cancer research. We have developed and evaluated convolutional neural network (CNN) analysis pipelines to generate combined maps of cancer regions and tumor infiltrating lymphocytes (TILs) in routine diagnostic breast cancer whole slide tissue images (WSIs). We produce interactive whole slide maps that provide 1) insight about the structural patterns and spatial distribution of lymphocytic infiltrates and 2) facilitate improved quantification of TILs. We evaluated both tumor and TIL analyses using three CNN networks - Resnet-34, VGG16 and Inception v4, and demonstrated that the results compared favorably to those obtained by what believe are the best published methods. We have produced open-source tools and generated a public dataset consisting of tumor/TIL maps for 1,015 TCGA breast cancer images. We also present a customized web-based interface that enables easy visualization and interactive exploration of high-resolution combined Tumor-TIL maps for 1,015TCGA invasive breast cancer cases that can be downloaded for further downstream analyses.
Label Super Resolution with Inter-Instance Loss
Apr 09, 2019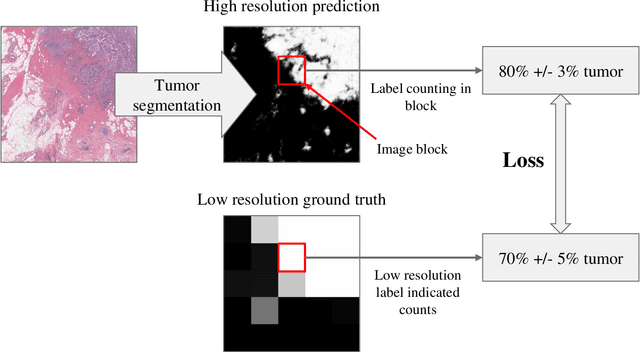
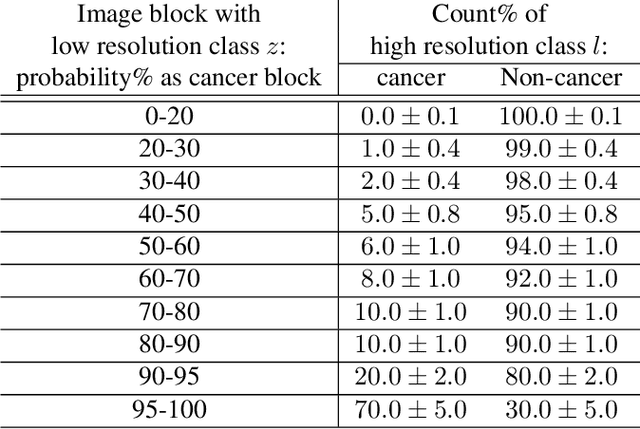
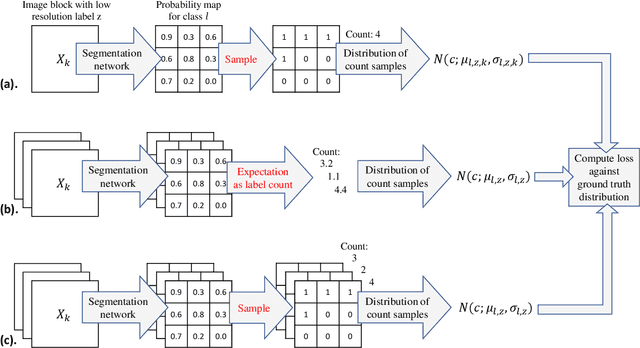
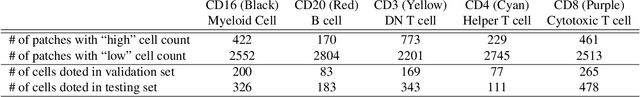
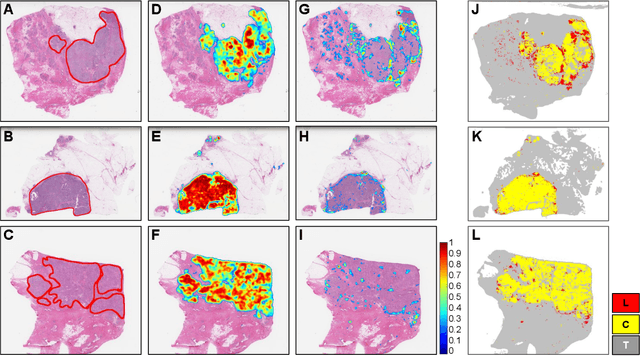
Abstract:For the task of semantic segmentation, high-resolution (pixel-level) ground truth is very expensive to collect, especially for high resolution images such as gigapixel pathology images. On the other hand, collecting low resolution labels (labels for a block of pixels) for these high resolution images is much more cost efficient. Conventional methods trained on these low-resolution labels are only capable of giving low-resolution predictions. The existing state-of-the-art label super resolution (LSR) method is capable of predicting high resolution labels, using only low-resolution supervision, given the joint distribution between low resolution and high resolution labels. However, it does not consider the inter-instance variance which is crucial in the ideal mathematical formulation. In this work, we propose a novel loss function modeling the inter-instance variance. We test our method on two real world applications: cell detection in multiplex immunohistochemistry (IHC) images, and infiltrating breast cancer region segmentation in histopathology slides. Experimental results show the effectiveness of our method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge