Shroyer Kenneth
Learning from Thresholds: Fully Automated Classification of Tumor Infiltrating Lymphocytes for Multiple Cancer Types
Jul 09, 2019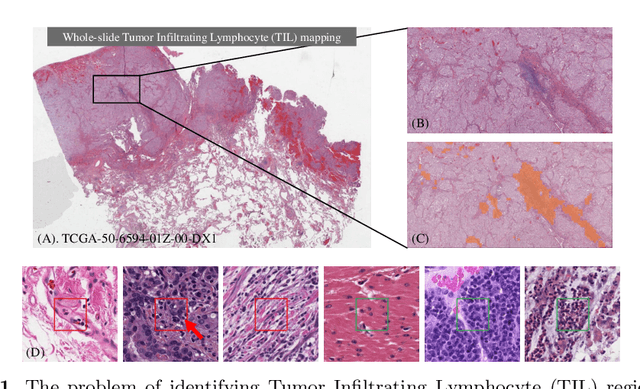

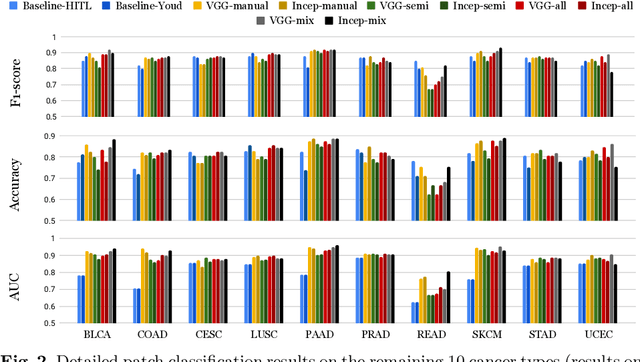
Abstract:Deep learning classifiers for characterization of whole slide tissue morphology require large volumes of annotated data to learn variations across different tissue and cancer types. As is well known, manual generation of digital pathology training data is time consuming and expensive. In this paper, we propose a semi-automated method for annotating a group of similar instances at once, instead of collecting only per-instance manual annotations. This allows for a much larger training set, that reflects visual variability across multiple cancer types and thus training of a single network which can be automatically applied to each cancer type without human adjustment. We apply our method to the important task of classifying Tumor Infiltrating Lymphocytes (TILs) in H&E images. Prior approaches were trained for individual cancer types, with smaller training sets and human-in-the-loop threshold adjustment. We utilize these thresholded results as large scale "semi-automatic" annotations. Combined with existing manual annotations, our trained deep networks are able to automatically produce better TIL prediction results in 12 cancer types, compared to the human-in-the-loop approach.
Label Super Resolution with Inter-Instance Loss
Apr 09, 2019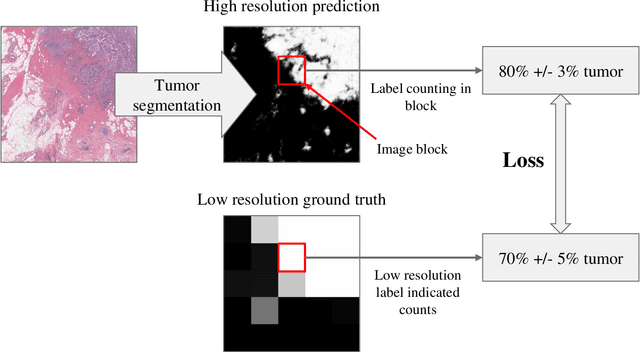
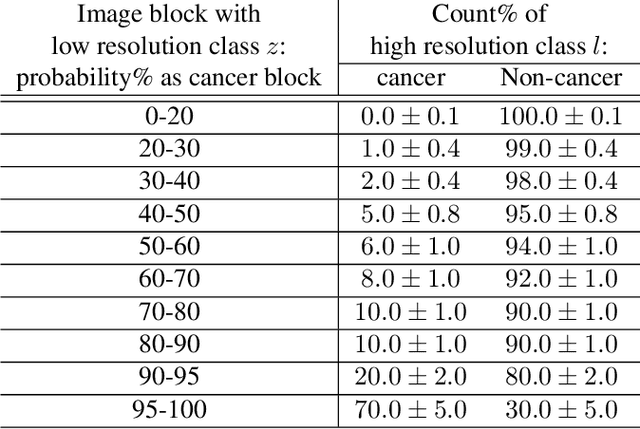
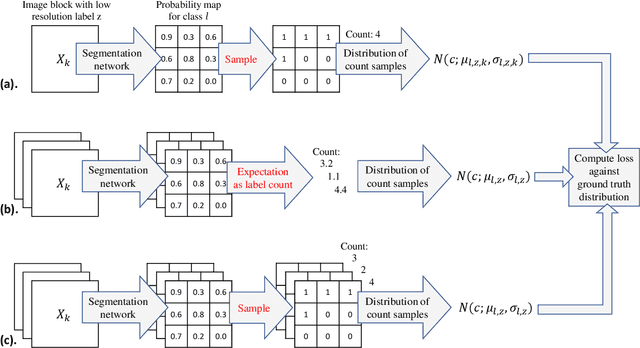
Abstract:For the task of semantic segmentation, high-resolution (pixel-level) ground truth is very expensive to collect, especially for high resolution images such as gigapixel pathology images. On the other hand, collecting low resolution labels (labels for a block of pixels) for these high resolution images is much more cost efficient. Conventional methods trained on these low-resolution labels are only capable of giving low-resolution predictions. The existing state-of-the-art label super resolution (LSR) method is capable of predicting high resolution labels, using only low-resolution supervision, given the joint distribution between low resolution and high resolution labels. However, it does not consider the inter-instance variance which is crucial in the ideal mathematical formulation. In this work, we propose a novel loss function modeling the inter-instance variance. We test our method on two real world applications: cell detection in multiplex immunohistochemistry (IHC) images, and infiltrating breast cancer region segmentation in histopathology slides. Experimental results show the effectiveness of our method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge