Talha Qaiser
A Novel Approach to Linking Histology Images with DNA Methylation
Apr 07, 2025Abstract:DNA methylation is an epigenetic mechanism that regulates gene expression by adding methyl groups to DNA. Abnormal methylation patterns can disrupt gene expression and have been linked to cancer development. To quantify DNA methylation, specialized assays are typically used. However, these assays are often costly and have lengthy processing times, which limits their widespread availability in routine clinical practice. In contrast, whole slide images (WSIs) for the majority of cancer patients can be more readily available. As such, given the ready availability of WSIs, there is a compelling need to explore the potential relationship between WSIs and DNA methylation patterns. To address this, we propose an end-to-end graph neural network based weakly supervised learning framework to predict the methylation state of gene groups exhibiting coherent patterns across samples. Using data from three cohorts from The Cancer Genome Atlas (TCGA) - TCGA-LGG (Brain Lower Grade Glioma), TCGA-GBM (Glioblastoma Multiforme) ($n$=729) and TCGA-KIRC (Kidney Renal Clear Cell Carcinoma) ($n$=511) - we demonstrate that the proposed approach achieves significantly higher AUROC scores than the state-of-the-art (SOTA) methods, by more than $20\%$. We conduct gene set enrichment analyses on the gene groups and show that majority of the gene groups are significantly enriched in important hallmarks and pathways. We also generate spatially enriched heatmaps to further investigate links between histological patterns and DNA methylation states. To the best of our knowledge, this is the first study that explores association of spatially resolved histological patterns with gene group methylation states across multiple cancer types using weakly supervised deep learning.
Stain-Invariant Representation for Tissue Classification in Histology Images
Nov 21, 2024
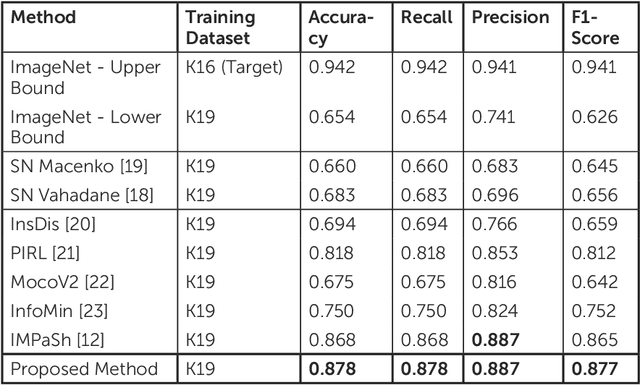
Abstract:The process of digitising histology slides involves multiple factors that can affect a whole slide image's (WSI) final appearance, including the staining protocol, scanner, and tissue type. This variability constitutes a domain shift and results in significant problems when training and testing deep learning (DL) algorithms in multi-cohort settings. As such, developing robust and generalisable DL models in computational pathology (CPath) remains an open challenge. In this regard, we propose a framework that generates stain-augmented versions of the training images using stain matrix perturbation. Thereafter, we employed a stain regularisation loss to enforce consistency between the feature representations of the source and augmented images. Doing so encourages the model to learn stain-invariant and, consequently, domain-invariant feature representations. We evaluate the performance of the proposed model on cross-domain multi-class tissue type classification of colorectal cancer images and have achieved improved performance compared to other state-of-the-art methods.
Can Medical Vision-Language Pre-training Succeed with Purely Synthetic Data?
Oct 17, 2024



Abstract:Medical Vision-Language Pre-training (MedVLP) has made significant progress in enabling zero-shot tasks for medical image understanding. However, training MedVLP models typically requires large-scale datasets with paired, high-quality image-text data, which are scarce in the medical domain. Recent advancements in Large Language Models (LLMs) and diffusion models have made it possible to generate large-scale synthetic image-text pairs. This raises the question: *Can MedVLP succeed using purely synthetic data?* To address this, we use off-the-shelf generative models to create synthetic radiology reports and paired Chest X-ray (CXR) images, and propose an automated pipeline to build a diverse, high-quality synthetic dataset, enabling a rigorous study that isolates model and training settings, focusing entirely from the data perspective. Our results show that MedVLP models trained *exclusively on synthetic data* outperform those trained on real data by **3.8%** in averaged AUC on zero-shot classification. Moreover, using a combination of synthetic and real data leads to a further improvement of **9.07%**. Additionally, MedVLP models trained on synthetic or mixed data consistently outperform those trained on real data in zero-shot grounding, as well as in fine-tuned classification and segmentation tasks. Our analysis suggests MedVLP trained on well-designed synthetic data can outperform models trained on real datasets, which may be limited by low-quality samples and long-tailed distributions.
Unsupervised Mutual Transformer Learning for Multi-Gigapixel Whole Slide Image Classification
May 03, 2023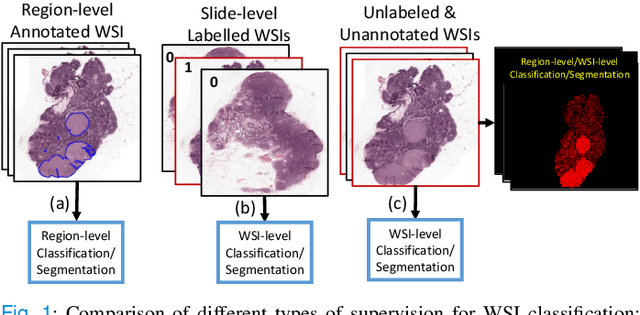
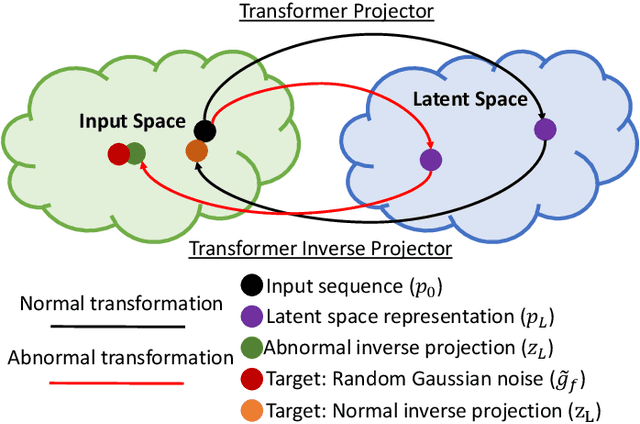
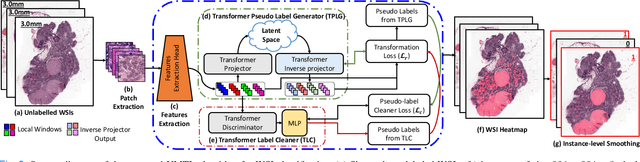
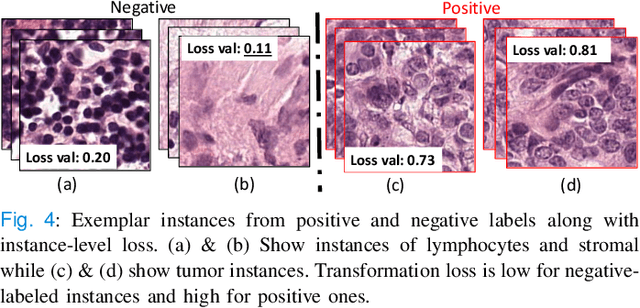
Abstract:Classification of gigapixel Whole Slide Images (WSIs) is an important prediction task in the emerging area of computational pathology. There has been a surge of research in deep learning models for WSI classification with clinical applications such as cancer detection or prediction of molecular mutations from WSIs. Most methods require expensive and labor-intensive manual annotations by expert pathologists. Weakly supervised Multiple Instance Learning (MIL) methods have recently demonstrated excellent performance; however, they still require large slide-level labeled training datasets that need a careful inspection of each slide by an expert pathologist. In this work, we propose a fully unsupervised WSI classification algorithm based on mutual transformer learning. Instances from gigapixel WSI (i.e., image patches) are transformed into a latent space and then inverse-transformed to the original space. Using the transformation loss, pseudo-labels are generated and cleaned using a transformer label-cleaner. The proposed transformer-based pseudo-label generation and cleaning modules mutually train each other iteratively in an unsupervised manner. A discriminative learning mechanism is introduced to improve normal versus cancerous instance labeling. In addition to unsupervised classification, we demonstrate the effectiveness of the proposed framework for weak supervision for cancer subtype classification as downstream analysis. Extensive experiments on four publicly available datasets show excellent performance compared to the state-of-the-art methods. We intend to make the source code of our algorithm publicly available soon.
Interpretable histopathology-based prediction of disease relevant features in Inflammatory Bowel Disease biopsies using weakly-supervised deep learning
Mar 20, 2023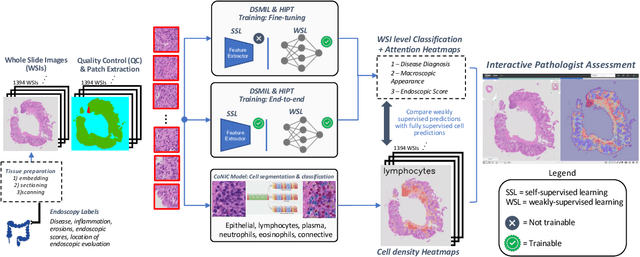
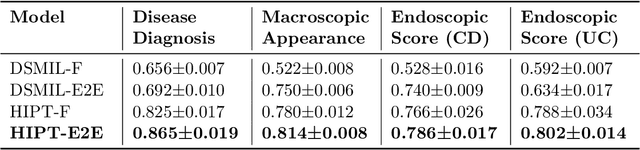
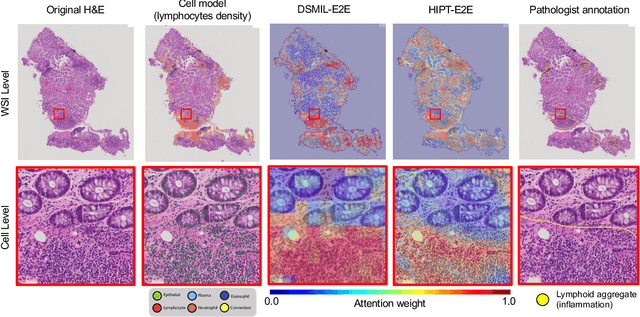
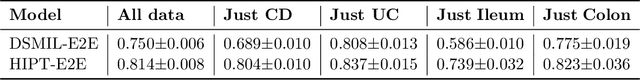
Abstract:Crohn's Disease (CD) and Ulcerative Colitis (UC) are the two main Inflammatory Bowel Disease (IBD) types. We developed deep learning models to identify histological disease features for both CD and UC using only endoscopic labels. We explored fine-tuning and end-to-end training of two state-of-the-art self-supervised models for predicting three different endoscopic categories (i) CD vs UC (AUC=0.87), (ii) normal vs lesional (AUC=0.81), (iii) low vs high disease severity score (AUC=0.80). We produced visual attention maps to interpret what the models learned and validated them with the support of a pathologist, where we observed a strong association between the models' predictions and histopathological inflammatory features of the disease. Additionally, we identified several cases where the model incorrectly predicted normal samples as lesional but were correct on the microscopic level when reviewed by the pathologist. This tendency of histological presentation to be more severe than endoscopic presentation was previously published in the literature. In parallel, we utilised a model trained on the Colon Nuclei Identification and Counting (CoNIC) dataset to predict and explore 6 cell populations. We observed correlation between areas enriched with the predicted immune cells in biopsies and the pathologist's feedback on the attention maps. Finally, we identified several cell level features indicative of disease severity in CD and UC. These models can enhance our understanding about the pathology behind IBD and can shape our strategies for patient stratification in clinical trials.
Mimicking a Pathologist: Dual Attention Model for Scoring of Gigapixel Histology Images
Feb 19, 2023


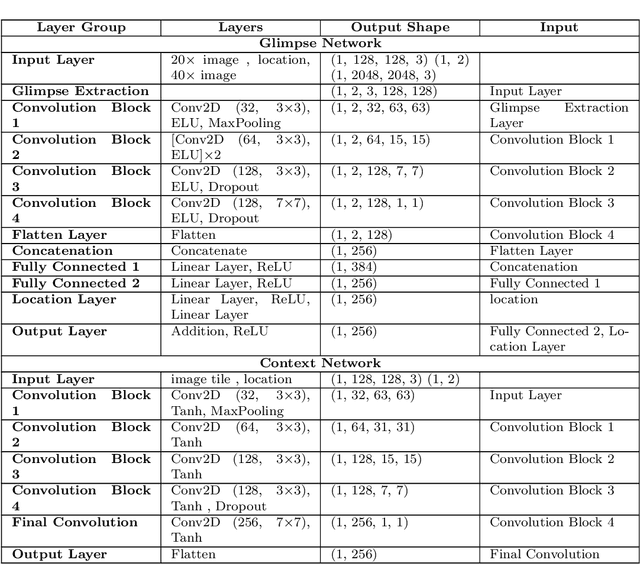
Abstract:Some major challenges associated with the automated processing of whole slide images (WSIs) includes their sheer size, different magnification levels and high resolution. Utilizing these images directly in AI frameworks is computationally expensive due to memory constraints, while downsampling WSIs incurs information loss and splitting WSIs into tiles and patches results in loss of important contextual information. We propose a novel dual attention approach, consisting of two main components, to mimic visual examination by a pathologist. The first component is a soft attention model which takes as input a high-level view of the WSI to determine various regions of interest. We employ a custom sampling method to extract diverse and spatially distinct image tiles from selected high attention areas. The second component is a hard attention classification model, which further extracts a sequence of multi-resolution glimpses from each tile for classification. Since hard attention is non-differentiable, we train this component using reinforcement learning and predict the location of glimpses without processing all patches of a given tile, thereby aligning with pathologist's way of diagnosis. We train our components both separately and in an end-to-end fashion using a joint loss function to demonstrate the efficacy of our proposed model. We employ our proposed model on two different IHC use cases: HER2 prediction on breast cancer and prediction of Intact/Loss status of two MMR biomarkers, for colorectal cancer. We show that the proposed model achieves accuracy comparable to state-of-the-art methods while only processing a small fraction of the WSI at highest magnification.
Consistency Regularisation in Varying Contexts and Feature Perturbations for Semi-Supervised Semantic Segmentation of Histology Images
Feb 11, 2023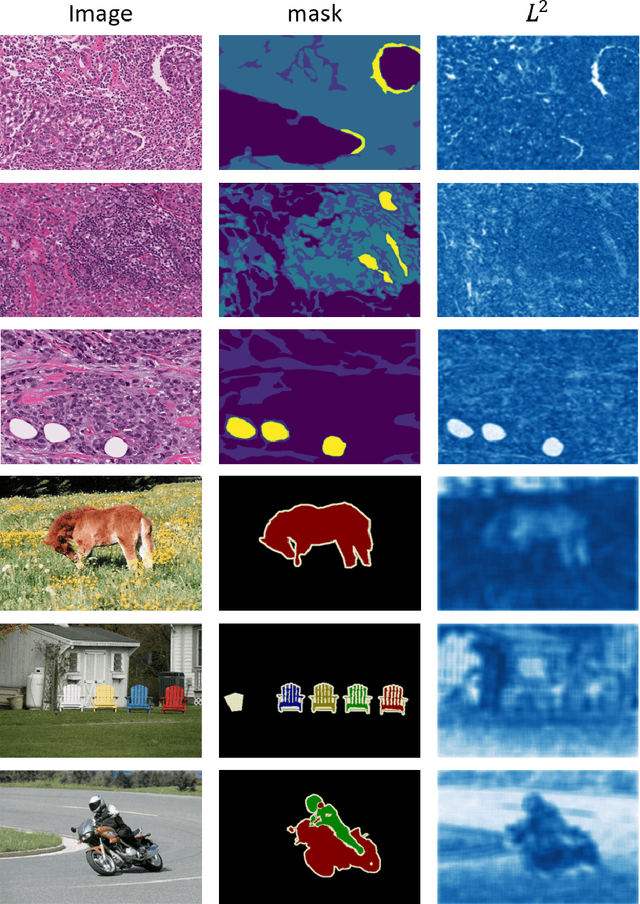
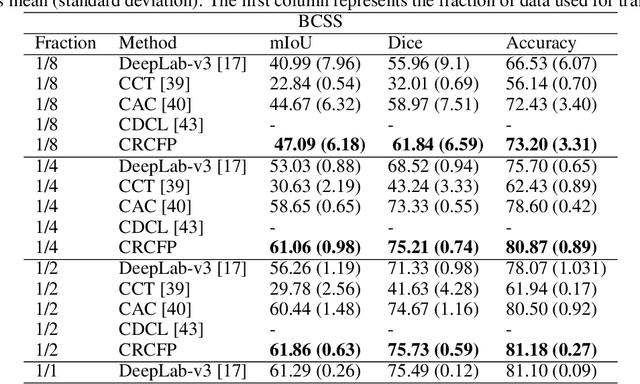
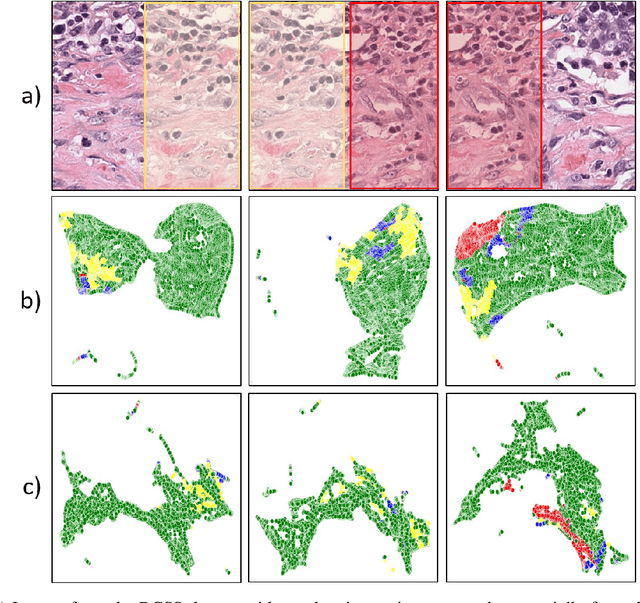
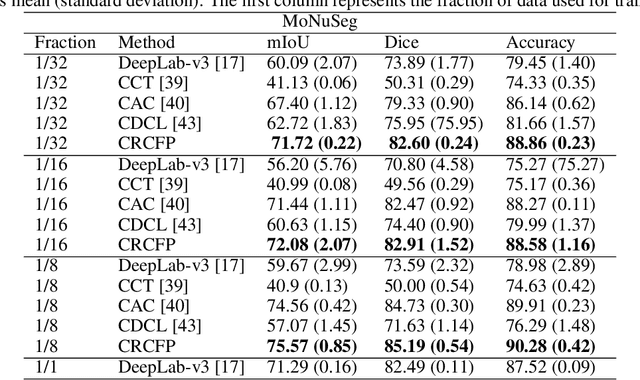
Abstract:Semantic segmentation of various tissue and nuclei types in histology images is fundamental to many downstream tasks in the area of computational pathology (CPath). In recent years, Deep Learning (DL) methods have been shown to perform well on segmentation tasks but DL methods generally require a large amount of pixel-wise annotated data. Pixel-wise annotation sometimes requires expert's knowledge and time which is laborious and costly to obtain. In this paper, we present a consistency based semi-supervised learning (SSL) approach that can help mitigate this challenge by exploiting a large amount of unlabelled data for model training thus alleviating the need for a large annotated dataset. However, SSL models might also be susceptible to changing context and features perturbations exhibiting poor generalisation due to the limited training data. We propose an SSL method that learns robust features from both labelled and unlabelled images by enforcing consistency against varying contexts and feature perturbations. The proposed method incorporates context-aware consistency by contrasting pairs of overlapping images in a pixel-wise manner from changing contexts resulting in robust and context invariant features. We show that cross-consistency training makes the encoder features invariant to different perturbations and improves the prediction confidence. Finally, entropy minimisation is employed to further boost the confidence of the final prediction maps from unlabelled data. We conduct an extensive set of experiments on two publicly available large datasets (BCSS and MoNuSeg) and show superior performance compared to the state-of-the-art methods.
Multiple Instance Learning with Auxiliary Task Weighting for Multiple Myeloma Classification
Jul 16, 2021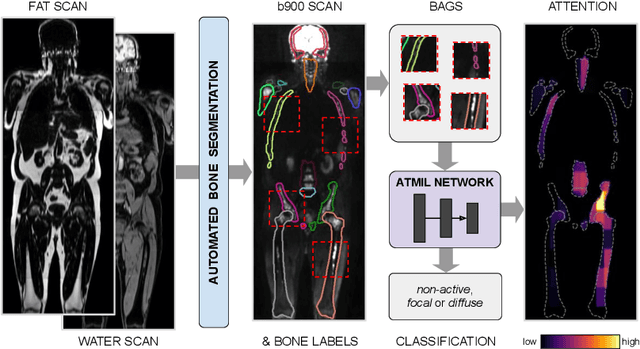
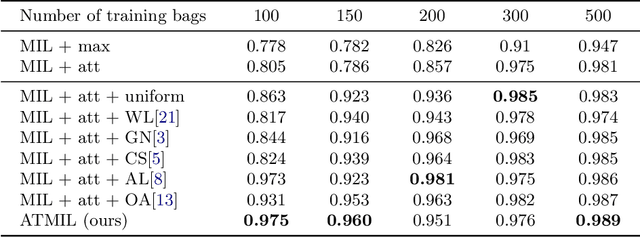
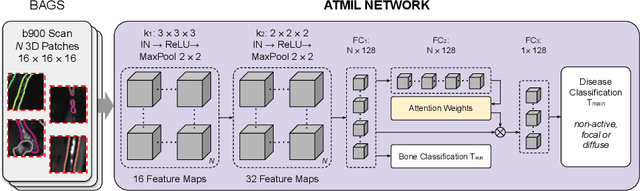
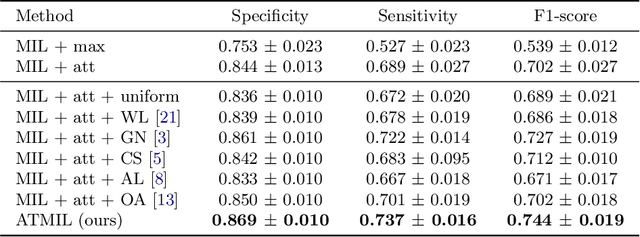
Abstract:Whole body magnetic resonance imaging (WB-MRI) is the recommended modality for diagnosis of multiple myeloma (MM). WB-MRI is used to detect sites of disease across the entire skeletal system, but it requires significant expertise and is time-consuming to report due to the great number of images. To aid radiological reading, we propose an auxiliary task-based multiple instance learning approach (ATMIL) for MM classification with the ability to localize sites of disease. This approach is appealing as it only requires patient-level annotations where an attention mechanism is used to identify local regions with active disease. We borrow ideas from multi-task learning and define an auxiliary task with adaptive reweighting to support and improve learning efficiency in the presence of data scarcity. We validate our approach on both synthetic and real multi-center clinical data. We show that the MIL attention module provides a mechanism to localize bone regions while the adaptive reweighting of the auxiliary task considerably improves the performance.
Classification of COVID-19 via Homology of CT-SCAN
Feb 21, 2021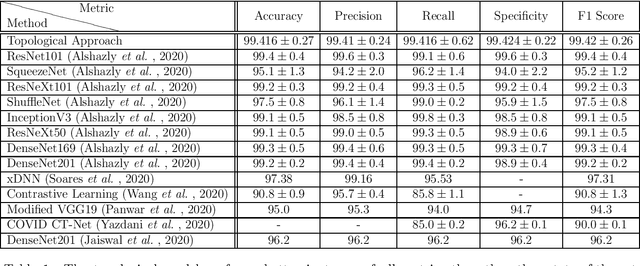
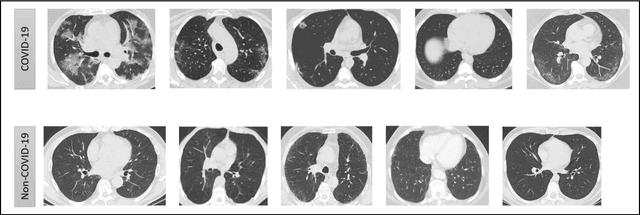
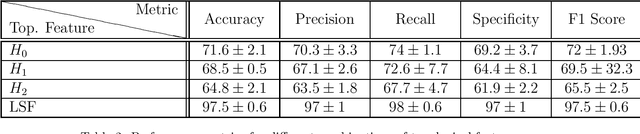
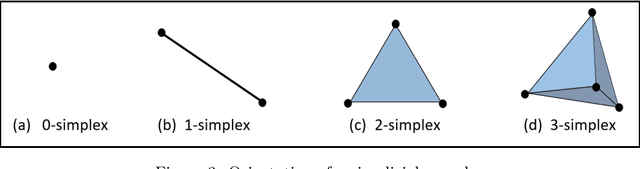
Abstract:In this worldwide spread of SARS-CoV-2 (COVID-19) infection, it is of utmost importance to detect the disease at an early stage especially in the hot spots of this epidemic. There are more than 110 Million infected cases on the globe, sofar. Due to its promptness and effective results computed tomography (CT)-scan image is preferred to the reverse-transcription polymerase chain reaction (RT-PCR). Early detection and isolation of the patient is the only possible way of controlling the spread of the disease. Automated analysis of CT-Scans can provide enormous support in this process. In this article, We propose a novel approach to detect SARS-CoV-2 using CT-scan images. Our method is based on a very intuitive and natural idea of analyzing shapes, an attempt to mimic a professional medic. We mainly trace SARS-CoV-2 features by quantifying their topological properties. We primarily use a tool called persistent homology, from Topological Data Analysis (TDA), to compute these topological properties. We train and test our model on the "SARS-CoV-2 CT-scan dataset" \citep{soares2020sars}, an open-source dataset, containing 2,481 CT-scans of normal and COVID-19 patients. Our model yielded an overall benchmark F1 score of $99.42\% $, accuracy $99.416\%$, precision $99.41\%$, and recall $99.42\%$. The TDA techniques have great potential that can be utilized for efficient and prompt detection of COVID-19. The immense potential of TDA may be exploited in clinics for rapid and safe detection of COVID-19 globally, in particular in the low and middle-income countries where RT-PCR labs and/or kits are in a serious crisis.
HydraMix-Net: A Deep Multi-task Semi-supervised Learning Approach for Cell Detection and Classification
Aug 11, 2020Abstract:Semi-supervised techniques have removed the barriers of large scale labelled set by exploiting unlabelled data to improve the performance of a model. In this paper, we propose a semi-supervised deep multi-task classification and localization approach HydraMix-Net in the field of medical imagining where labelling is time consuming and costly. Firstly, the pseudo labels are generated using the model's prediction on the augmented set of unlabelled image with averaging. The high entropy predictions are further sharpened to reduced the entropy and are then mixed with the labelled set for training. The model is trained in multi-task learning manner with noise tolerant joint loss for classification localization and achieves better performance when given limited data in contrast to a simple deep model. On DLBCL data it achieves 80\% accuracy in contrast to simple CNN achieving 70\% accuracy when given only 100 labelled examples.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge