Azam Hamidinekoo
Interpretable histopathology-based prediction of disease relevant features in Inflammatory Bowel Disease biopsies using weakly-supervised deep learning
Mar 20, 2023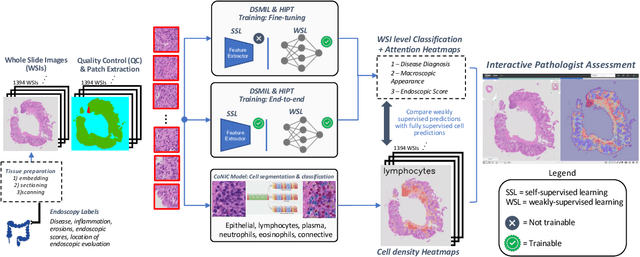
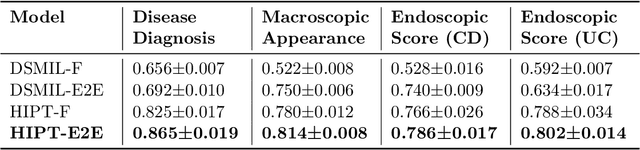
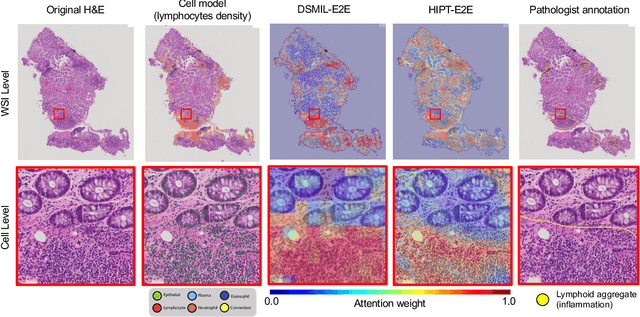
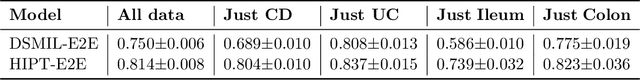
Abstract:Crohn's Disease (CD) and Ulcerative Colitis (UC) are the two main Inflammatory Bowel Disease (IBD) types. We developed deep learning models to identify histological disease features for both CD and UC using only endoscopic labels. We explored fine-tuning and end-to-end training of two state-of-the-art self-supervised models for predicting three different endoscopic categories (i) CD vs UC (AUC=0.87), (ii) normal vs lesional (AUC=0.81), (iii) low vs high disease severity score (AUC=0.80). We produced visual attention maps to interpret what the models learned and validated them with the support of a pathologist, where we observed a strong association between the models' predictions and histopathological inflammatory features of the disease. Additionally, we identified several cases where the model incorrectly predicted normal samples as lesional but were correct on the microscopic level when reviewed by the pathologist. This tendency of histological presentation to be more severe than endoscopic presentation was previously published in the literature. In parallel, we utilised a model trained on the Colon Nuclei Identification and Counting (CoNIC) dataset to predict and explore 6 cell populations. We observed correlation between areas enriched with the predicted immune cells in biopsies and the pathologist's feedback on the attention maps. Finally, we identified several cell level features indicative of disease severity in CD and UC. These models can enhance our understanding about the pathology behind IBD and can shape our strategies for patient stratification in clinical trials.
Glioma Classification Using Multimodal Radiology and Histology Data
Nov 10, 2020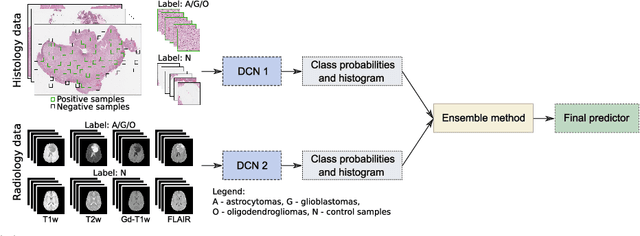
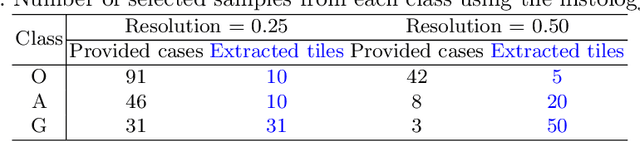
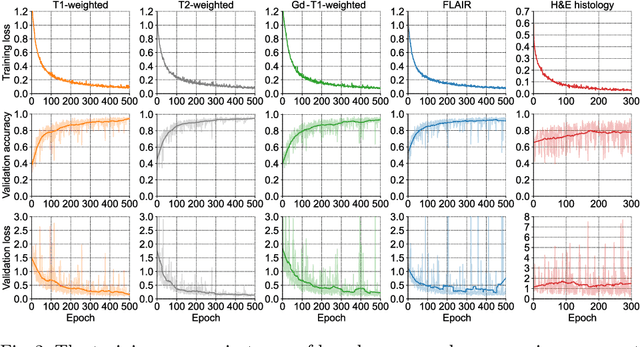
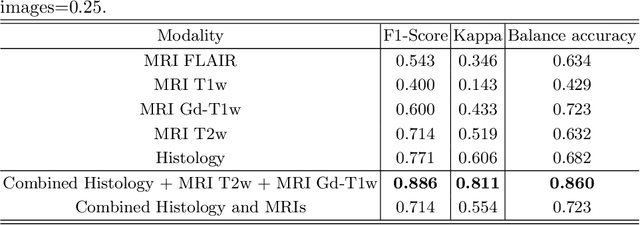
Abstract:Gliomas are brain tumours with a high mortality rate. There are various grades and sub-types of this tumour, and the treatment procedure varies accordingly. Clinicians and oncologists diagnose and categorise these tumours based on visual inspection of radiology and histology data. However, this process can be time-consuming and subjective. The computer-assisted methods can help clinicians to make better and faster decisions. In this paper, we propose a pipeline for automatic classification of gliomas into three sub-types: oligodendroglioma, astrocytoma, and glioblastoma, using both radiology and histopathology images. The proposed approach implements distinct classification models for radiographic and histologic modalities and combines them through an ensemble method. The classification algorithm initially carries out tile-level (for histology) and slice-level (for radiology) classification via a deep learning method, then tile/slice-level latent features are combined for a whole-slide and whole-volume sub-type prediction. The classification algorithm was evaluated using the data set provided in the CPM-RadPath 2020 challenge. The proposed pipeline achieved the F1-Score of 0.886, Cohen's Kappa score of 0.811 and Balance accuracy of 0.860. The ability of the proposed model for end-to-end learning of diverse features enables it to give a comparable prediction of glioma tumour sub-types.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge