Junmei Cairns
Interpretable histopathology-based prediction of disease relevant features in Inflammatory Bowel Disease biopsies using weakly-supervised deep learning
Mar 20, 2023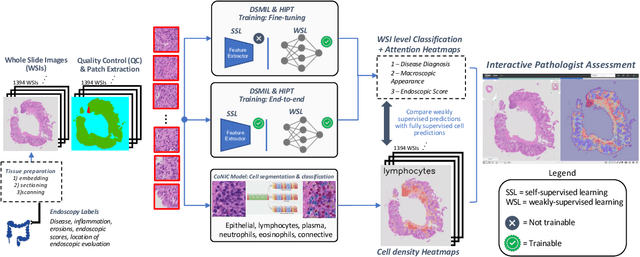
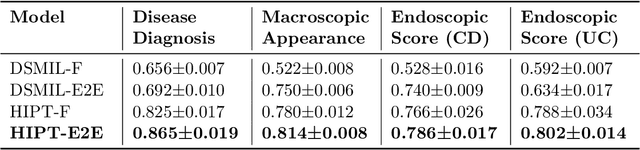
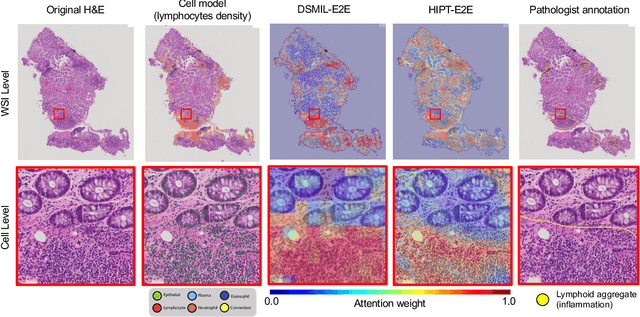
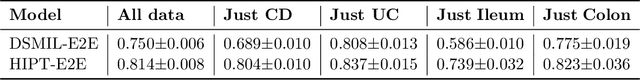
Abstract:Crohn's Disease (CD) and Ulcerative Colitis (UC) are the two main Inflammatory Bowel Disease (IBD) types. We developed deep learning models to identify histological disease features for both CD and UC using only endoscopic labels. We explored fine-tuning and end-to-end training of two state-of-the-art self-supervised models for predicting three different endoscopic categories (i) CD vs UC (AUC=0.87), (ii) normal vs lesional (AUC=0.81), (iii) low vs high disease severity score (AUC=0.80). We produced visual attention maps to interpret what the models learned and validated them with the support of a pathologist, where we observed a strong association between the models' predictions and histopathological inflammatory features of the disease. Additionally, we identified several cases where the model incorrectly predicted normal samples as lesional but were correct on the microscopic level when reviewed by the pathologist. This tendency of histological presentation to be more severe than endoscopic presentation was previously published in the literature. In parallel, we utilised a model trained on the Colon Nuclei Identification and Counting (CoNIC) dataset to predict and explore 6 cell populations. We observed correlation between areas enriched with the predicted immune cells in biopsies and the pathologist's feedback on the attention maps. Finally, we identified several cell level features indicative of disease severity in CD and UC. These models can enhance our understanding about the pathology behind IBD and can shape our strategies for patient stratification in clinical trials.
Considerations of automated machine learning in clinical metabolic profiling: Altered homocysteine plasma concentration associated with metformin exposure
Oct 09, 2017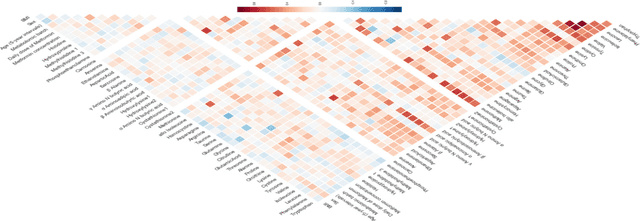
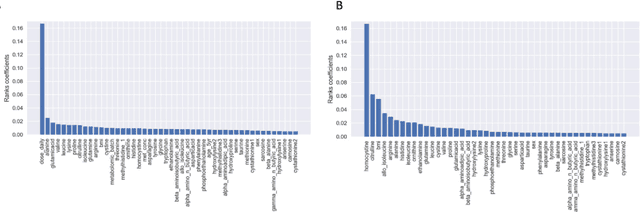
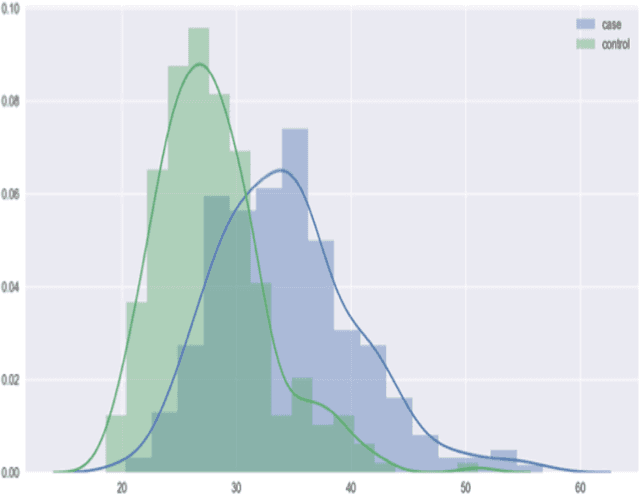
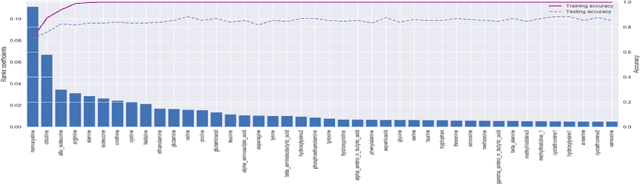
Abstract:With the maturation of metabolomics science and proliferation of biobanks, clinical metabolic profiling is an increasingly opportunistic frontier for advancing translational clinical research. Automated Machine Learning (AutoML) approaches provide exciting opportunity to guide feature selection in agnostic metabolic profiling endeavors, where potentially thousands of independent data points must be evaluated. In previous research, AutoML using high-dimensional data of varying types has been demonstrably robust, outperforming traditional approaches. However, considerations for application in clinical metabolic profiling remain to be evaluated. Particularly, regarding the robustness of AutoML to identify and adjust for common clinical confounders. In this study, we present a focused case study regarding AutoML considerations for using the Tree-Based Optimization Tool (TPOT) in metabolic profiling of exposure to metformin in a biobank cohort. First, we propose a tandem rank-accuracy measure to guide agnostic feature selection and corresponding threshold determination in clinical metabolic profiling endeavors. Second, while AutoML, using default parameters, demonstrated potential to lack sensitivity to low-effect confounding clinical covariates, we demonstrated residual training and adjustment of metabolite features as an easily applicable approach to ensure AutoML adjustment for potential confounding characteristics. Finally, we present increased homocysteine with long-term exposure to metformin as a potentially novel, non-replicated metabolite association suggested by TPOT; an association not identified in parallel clinical metabolic profiling endeavors. While considerations are recommended, including adjustment approaches for clinical confounders, AutoML presents an exciting tool to enhance clinical metabolic profiling and advance translational research endeavors.
* Manuscript - containing supplementary information - accepted (9/15/2017) for publication within Pacific Symposium on Biocomputing 2018 <https://psb.stanford.edu/psb-online>. Original supplementary information includes an additional 6 pages of content (18 pages total) and 8 figures (13 figures total)
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge