Pedro J. Caraballo
A novel method for Causal Structure Discovery from EHR data, a demonstration on type-2 diabetes mellitus
Nov 11, 2020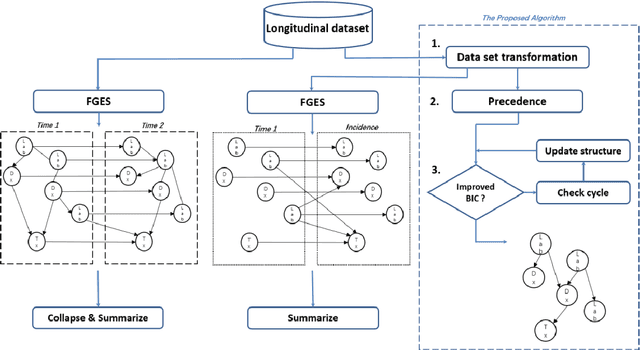
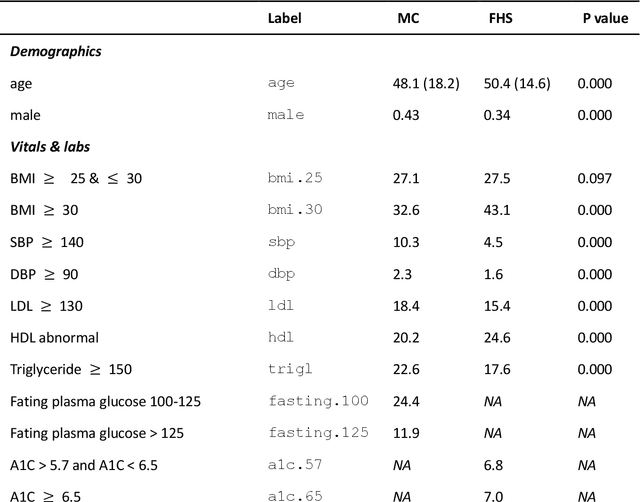
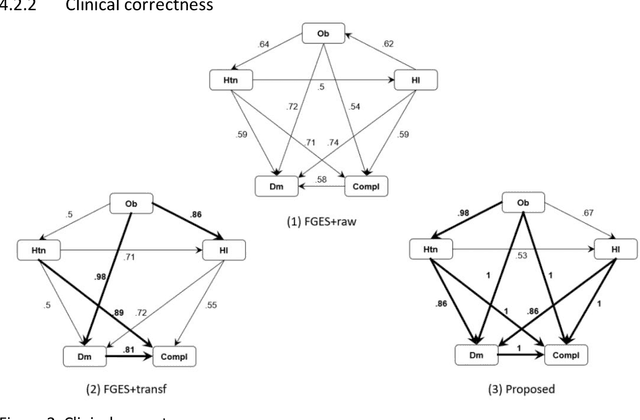
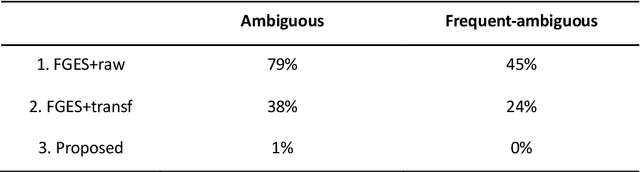
Abstract:Introduction: The discovery of causal mechanisms underlying diseases enables better diagnosis, prognosis and treatment selection. Clinical trials have been the gold standard for determining causality, but they are resource intensive, sometimes infeasible or unethical. Electronic Health Records (EHR) contain a wealth of real-world data that holds promise for the discovery of disease mechanisms, yet the existing causal structure discovery (CSD) methods fall short on leveraging them due to the special characteristics of the EHR data. We propose a new data transformation method and a novel CSD algorithm to overcome the challenges posed by these characteristics. Materials and methods: We demonstrated the proposed methods on an application to type-2 diabetes mellitus. We used a large EHR data set from Mayo Clinic to internally evaluate the proposed transformation and CSD methods and used another large data set from an independent health system, Fairview Health Services, as external validation. We compared the performance of our proposed method to Fast Greedy Equivalence Search (FGES), a state-of-the-art CSD method in terms of correctness, stability and completeness. We tested the generalizability of the proposed algorithm through external validation. Results and conclusions: The proposed method improved over the existing methods by successfully incorporating study design considerations, was robust in face of unreliable EHR timestamps and inferred causal effect directions more correctly and reliably. The proposed data transformation successfully improved the clinical correctness of the discovered graph and the consistency of edge orientation across bootstrap samples. It resulted in superior accuracy, stability, and completeness.
Considerations of automated machine learning in clinical metabolic profiling: Altered homocysteine plasma concentration associated with metformin exposure
Oct 09, 2017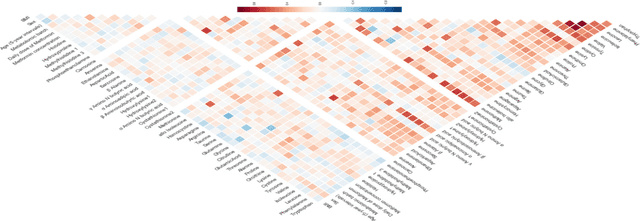
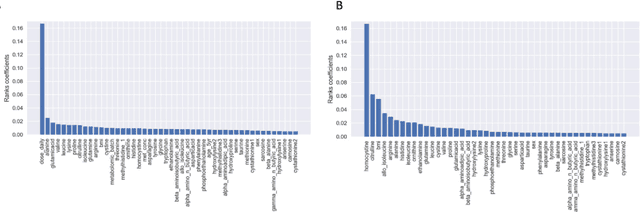
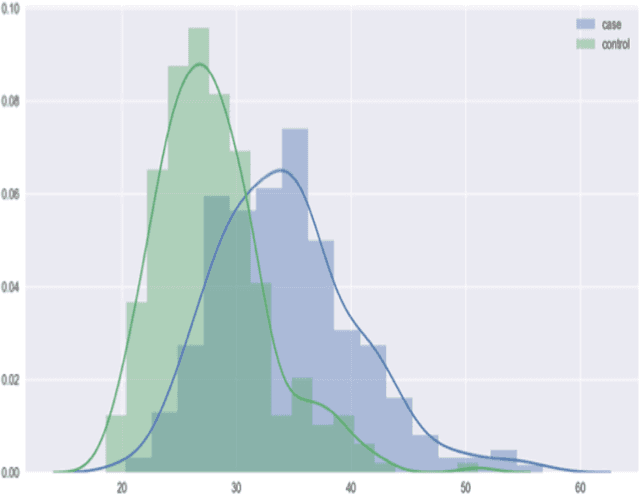
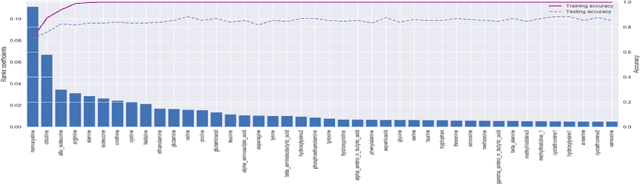
Abstract:With the maturation of metabolomics science and proliferation of biobanks, clinical metabolic profiling is an increasingly opportunistic frontier for advancing translational clinical research. Automated Machine Learning (AutoML) approaches provide exciting opportunity to guide feature selection in agnostic metabolic profiling endeavors, where potentially thousands of independent data points must be evaluated. In previous research, AutoML using high-dimensional data of varying types has been demonstrably robust, outperforming traditional approaches. However, considerations for application in clinical metabolic profiling remain to be evaluated. Particularly, regarding the robustness of AutoML to identify and adjust for common clinical confounders. In this study, we present a focused case study regarding AutoML considerations for using the Tree-Based Optimization Tool (TPOT) in metabolic profiling of exposure to metformin in a biobank cohort. First, we propose a tandem rank-accuracy measure to guide agnostic feature selection and corresponding threshold determination in clinical metabolic profiling endeavors. Second, while AutoML, using default parameters, demonstrated potential to lack sensitivity to low-effect confounding clinical covariates, we demonstrated residual training and adjustment of metabolite features as an easily applicable approach to ensure AutoML adjustment for potential confounding characteristics. Finally, we present increased homocysteine with long-term exposure to metformin as a potentially novel, non-replicated metabolite association suggested by TPOT; an association not identified in parallel clinical metabolic profiling endeavors. While considerations are recommended, including adjustment approaches for clinical confounders, AutoML presents an exciting tool to enhance clinical metabolic profiling and advance translational research endeavors.
* Manuscript - containing supplementary information - accepted (9/15/2017) for publication within Pacific Symposium on Biocomputing 2018 <https://psb.stanford.edu/psb-online>. Original supplementary information includes an additional 6 pages of content (18 pages total) and 8 figures (13 figures total)
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge