Dow-Mu Koh
Generalizing imaging biomarker repeatability studies using Bayesian inference: Applications in detecting heterogeneous treatment response in whole-body diffusion-weighted MRI of metastatic prostate cancer
May 14, 2025Abstract:The assessment of imaging biomarkers is critical for advancing precision medicine and improving disease characterization. Despite the availability of methods to derive disease heterogeneity metrics in imaging studies, a robust framework for evaluating measurement uncertainty remains underdeveloped. To address this gap, we propose a novel Bayesian framework to assess the precision of disease heterogeneity measures in biomarker studies. Our approach extends traditional methods for evaluating biomarker precision by providing greater flexibility in statistical assumptions and enabling the analysis of biomarkers beyond univariate or multivariate normally-distributed variables. Using Hamiltonian Monte Carlo sampling, the framework supports both, for example, normally-distributed and Dirichlet-Multinomial distributed variables, enabling the derivation of posterior distributions for biomarker parameters under diverse model assumptions. Designed to be broadly applicable across various imaging modalities and biomarker types, the framework builds a foundation for generalizing reproducible and objective biomarker evaluation. To demonstrate utility, we apply the framework to whole-body diffusion-weighted MRI (WBDWI) to assess heterogeneous therapeutic responses in metastatic bone disease. Specifically, we analyze data from two patient studies investigating treatments for metastatic castrate-resistant prostate cancer (mCRPC). Our results reveal an approximately 70% response rate among individual tumors across both studies, objectively characterizing differential responses to systemic therapies and validating the clinical relevance of the proposed methodology. This Bayesian framework provides a powerful tool for advancing biomarker research across diverse imaging-based studies while offering valuable insights into specific clinical applications, such as mCRPC treatment response.
Signal-based AI-driven software solution for automated quantification of metastatic bone disease and treatment response assessment using Whole-Body Diffusion-Weighted MRI (WB-DWI) biomarkers in Advanced Prostate Cancer
May 13, 2025Abstract:We developed an AI-driven software solution to quantify metastatic bone disease from WB-DWI scans. Core technologies include: (i) a weakly-supervised Residual U-Net model generating a skeleton probability map to isolate bone; (ii) a statistical framework for WB-DWI intensity normalisation, obtaining a signal-normalised b=900s/mm^2 (b900) image; and (iii) a shallow convolutional neural network that processes outputs from (i) and (ii) to generate a mask of suspected bone lesions, characterised by higher b900 signal intensity due to restricted water diffusion. This mask is applied to the gADC map to extract TDV and gADC statistics. We tested the tool using expert-defined metastatic bone disease delineations on 66 datasets, assessed repeatability of imaging biomarkers (N=10), and compared software-based response assessment with a construct reference standard based on clinical, laboratory and imaging assessments (N=118). Dice score between manual and automated delineations was 0.6 for lesions within pelvis and spine, with an average surface distance of 2mm. Relative differences for log-transformed TDV (log-TDV) and median gADC were below 9% and 5%, respectively. Repeatability analysis showed coefficients of variation of 4.57% for log-TDV and 3.54% for median gADC, with intraclass correlation coefficients above 0.9. The software achieved 80.5% accuracy, 84.3% sensitivity, and 85.7% specificity in assessing response to treatment compared to the construct reference standard. Computation time generating a mask averaged 90 seconds per scan. Our software enables reproducible TDV and gADC quantification from WB-DWI scans for monitoring metastatic bone disease response, thus providing potentially useful measurements for clinical decision-making in APC patients.
Deep Learning Framework with Multi-Head Dilated Encoders for Enhanced Segmentation of Cervical Cancer on Multiparametric Magnetic Resonance Imaging
Jun 19, 2023



Abstract:T2-weighted magnetic resonance imaging (MRI) and diffusion-weighted imaging (DWI) are essential components for cervical cancer diagnosis. However, combining these channels for training deep learning models are challenging due to misalignment of images. Here, we propose a novel multi-head framework that uses dilated convolutions and shared residual connections for separate encoding of multiparametric MRI images. We employ a residual U-Net model as a baseline, and perform a series of architectural experiments to evaluate the tumor segmentation performance based on multiparametric input channels and feature encoding configurations. All experiments were performed using a cohort including 207 patients with locally advanced cervical cancer. Our proposed multi-head model using separate dilated encoding for T2W MRI, and combined b1000 DWI and apparent diffusion coefficient (ADC) images achieved the best median Dice coefficient similarity (DSC) score, 0.823 (95% confidence interval (CI), 0.595-0.797), outperforming the conventional multi-channel model, DSC 0.788 (95% CI, 0.568-0.776), although the difference was not statistically significant (p>0.05). We investigated channel sensitivity using 3D GRAD-CAM and channel dropout, and highlighted the critical importance of T2W and ADC channels for accurate tumor segmentations. However, our results showed that b1000 DWI had a minor impact on overall segmentation performance. We demonstrated that the use of separate dilated feature extractors and independent contextual learning improved the model's ability to reduce the boundary effects and distortion of DWI, leading to improved segmentation performance. Our findings can have significant implications for the development of robust and generalizable models that can extend to other multi-modal segmentation applications.
Multiple Instance Learning with Auxiliary Task Weighting for Multiple Myeloma Classification
Jul 16, 2021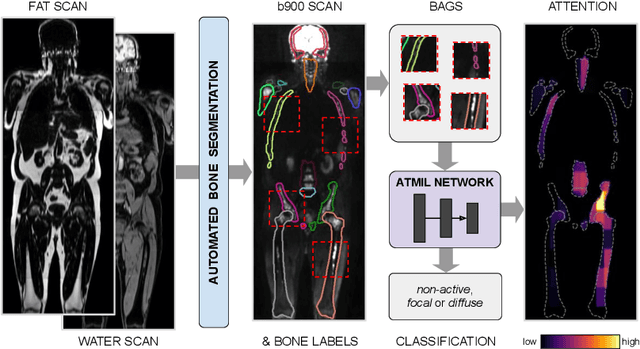
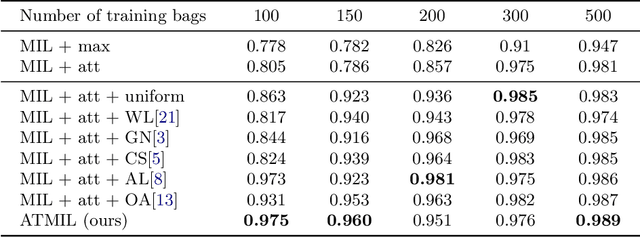
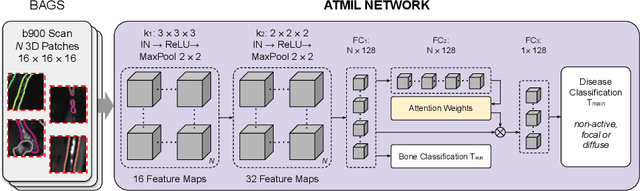
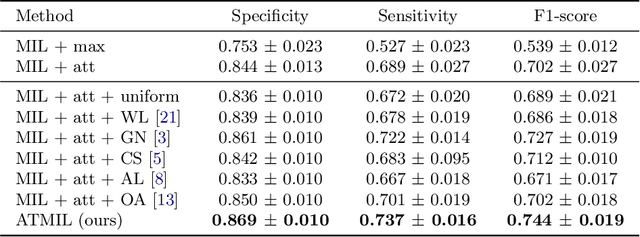
Abstract:Whole body magnetic resonance imaging (WB-MRI) is the recommended modality for diagnosis of multiple myeloma (MM). WB-MRI is used to detect sites of disease across the entire skeletal system, but it requires significant expertise and is time-consuming to report due to the great number of images. To aid radiological reading, we propose an auxiliary task-based multiple instance learning approach (ATMIL) for MM classification with the ability to localize sites of disease. This approach is appealing as it only requires patient-level annotations where an attention mechanism is used to identify local regions with active disease. We borrow ideas from multi-task learning and define an auxiliary task with adaptive reweighting to support and improve learning efficiency in the presence of data scarcity. We validate our approach on both synthetic and real multi-center clinical data. We show that the MIL attention module provides a mechanism to localize bone regions while the adaptive reweighting of the auxiliary task considerably improves the performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge