Gigin Lin
Deep Learning Framework with Multi-Head Dilated Encoders for Enhanced Segmentation of Cervical Cancer on Multiparametric Magnetic Resonance Imaging
Jun 19, 2023



Abstract:T2-weighted magnetic resonance imaging (MRI) and diffusion-weighted imaging (DWI) are essential components for cervical cancer diagnosis. However, combining these channels for training deep learning models are challenging due to misalignment of images. Here, we propose a novel multi-head framework that uses dilated convolutions and shared residual connections for separate encoding of multiparametric MRI images. We employ a residual U-Net model as a baseline, and perform a series of architectural experiments to evaluate the tumor segmentation performance based on multiparametric input channels and feature encoding configurations. All experiments were performed using a cohort including 207 patients with locally advanced cervical cancer. Our proposed multi-head model using separate dilated encoding for T2W MRI, and combined b1000 DWI and apparent diffusion coefficient (ADC) images achieved the best median Dice coefficient similarity (DSC) score, 0.823 (95% confidence interval (CI), 0.595-0.797), outperforming the conventional multi-channel model, DSC 0.788 (95% CI, 0.568-0.776), although the difference was not statistically significant (p>0.05). We investigated channel sensitivity using 3D GRAD-CAM and channel dropout, and highlighted the critical importance of T2W and ADC channels for accurate tumor segmentations. However, our results showed that b1000 DWI had a minor impact on overall segmentation performance. We demonstrated that the use of separate dilated feature extractors and independent contextual learning improved the model's ability to reduce the boundary effects and distortion of DWI, leading to improved segmentation performance. Our findings can have significant implications for the development of robust and generalizable models that can extend to other multi-modal segmentation applications.
Lesion Segmentation and RECIST Diameter Prediction via Click-driven Attention and Dual-path Connection
May 05, 2021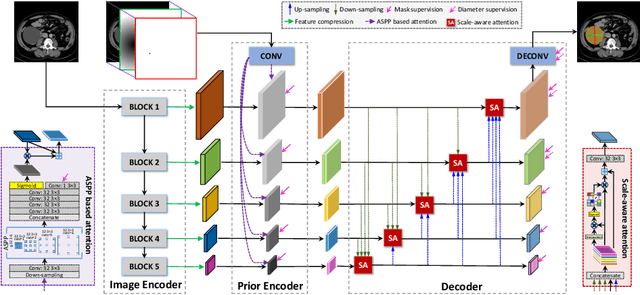
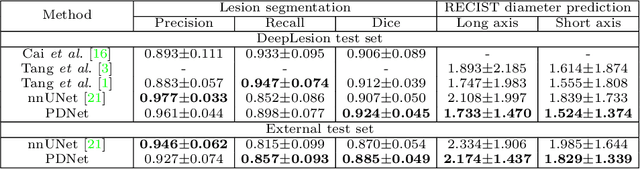
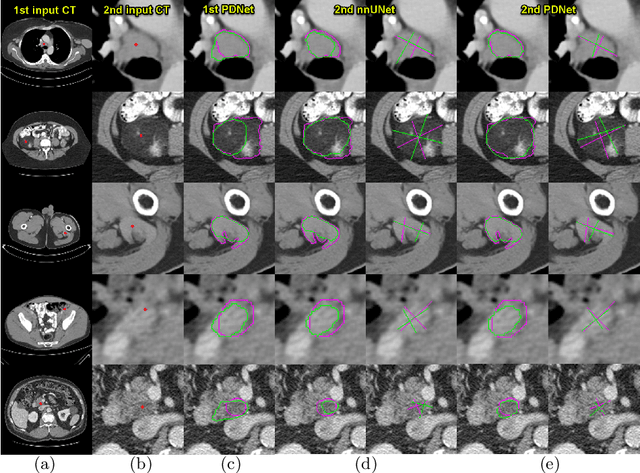
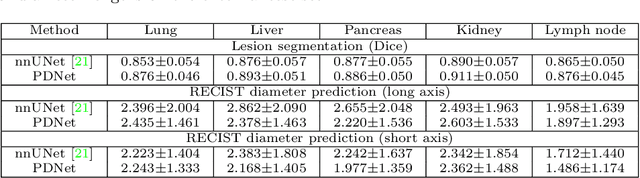
Abstract:Measuring lesion size is an important step to assess tumor growth and monitor disease progression and therapy response in oncology image analysis. Although it is tedious and highly time-consuming, radiologists have to work on this task by using RECIST criteria (Response Evaluation Criteria In Solid Tumors) routinely and manually. Even though lesion segmentation may be the more accurate and clinically more valuable means, physicians can not manually segment lesions as now since much more heavy laboring will be required. In this paper, we present a prior-guided dual-path network (PDNet) to segment common types of lesions throughout the whole body and predict their RECIST diameters accurately and automatically. Similar to [1], a click guidance from radiologists is the only requirement. There are two key characteristics in PDNet: 1) Learning lesion-specific attention matrices in parallel from the click prior information by the proposed prior encoder, named click-driven attention; 2) Aggregating the extracted multi-scale features comprehensively by introducing top-down and bottom-up connections in the proposed decoder, named dual-path connection. Experiments show the superiority of our proposed PDNet in lesion segmentation and RECIST diameter prediction using the DeepLesion dataset and an external test set. PDNet learns comprehensive and representative deep image features for our tasks and produces more accurate results on both lesion segmentation and RECIST diameter prediction.
Weakly-Supervised Universal Lesion Segmentation with Regional Level Set Loss
May 03, 2021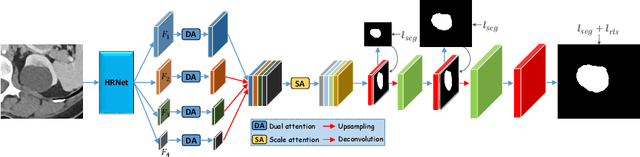
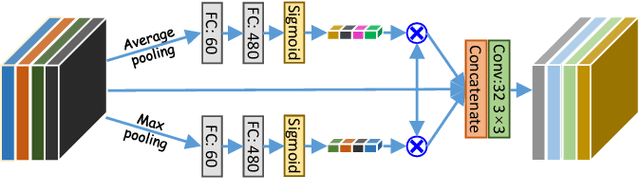
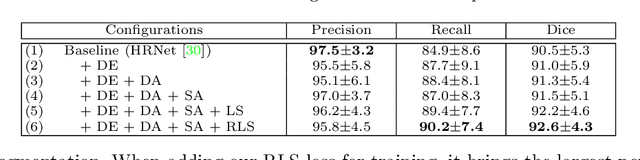
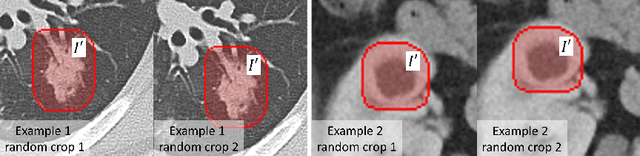
Abstract:Accurately segmenting a variety of clinically significant lesions from whole body computed tomography (CT) scans is a critical task on precision oncology imaging, denoted as universal lesion segmentation (ULS). Manual annotation is the current clinical practice, being highly time-consuming and inconsistent on tumor's longitudinal assessment. Effectively training an automatic segmentation model is desirable but relies heavily on a large number of pixel-wise labelled data. Existing weakly-supervised segmentation approaches often struggle with regions nearby the lesion boundaries. In this paper, we present a novel weakly-supervised universal lesion segmentation method by building an attention enhanced model based on the High-Resolution Network (HRNet), named AHRNet, and propose a regional level set (RLS) loss for optimizing lesion boundary delineation. AHRNet provides advanced high-resolution deep image features by involving a decoder, dual-attention and scale attention mechanisms, which are crucial to performing accurate lesion segmentation. RLS can optimize the model reliably and effectively in a weakly-supervised fashion, forcing the segmentation close to lesion boundary. Extensive experimental results demonstrate that our method achieves the best performance on the publicly large-scale DeepLesion dataset and a hold-out test set.
Deep Lesion Tracker: Monitoring Lesions in 4D Longitudinal Imaging Studies
Dec 09, 2020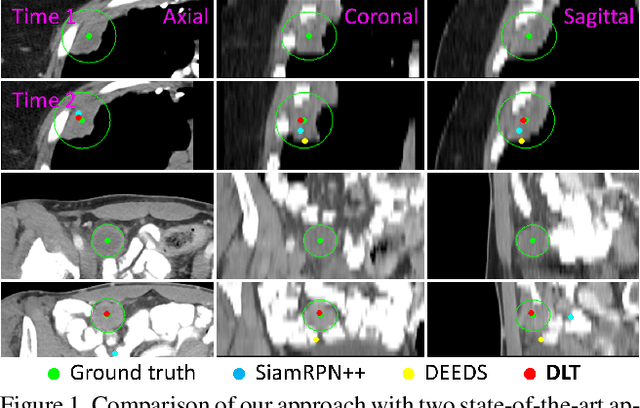
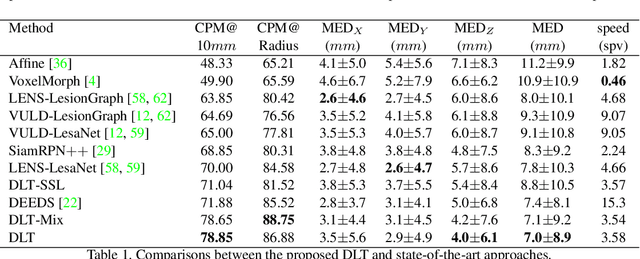
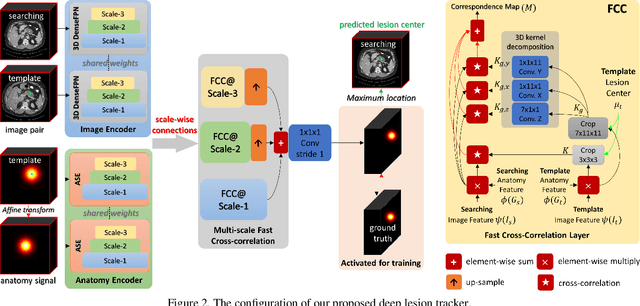
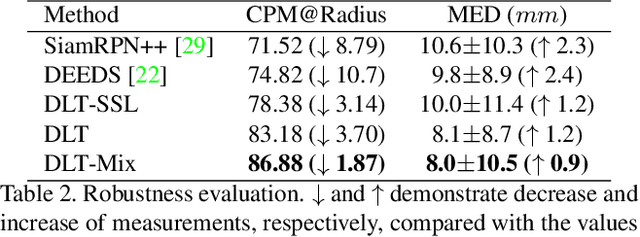
Abstract:Monitoring treatment response in longitudinal studies plays an important role in clinical practice. Accurately identifying lesions across serial imaging follow-up is the core to the monitoring procedure. Typically this incorporates both image and anatomical considerations. However, matching lesions manually is labor-intensive and time-consuming. In this work, we present deep lesion tracker (DLT), a deep learning approach that uses both appearance- and anatomical-based signals. To incorporate anatomical constraints, we propose an anatomical signal encoder, which prevents lesions being matched with visually similar but spurious regions. In addition, we present a new formulation for Siamese networks that avoids the heavy computational loads of 3D cross-correlation. To present our network with greater varieties of images, we also propose a self-supervised learning (SSL) strategy to train trackers with unpaired images, overcoming barriers to data collection. To train and evaluate our tracker, we introduce and release the first lesion tracking benchmark, consisting of 3891 lesion pairs from the public DeepLesion database. The proposed method, DLT, locates lesion centers with a mean error distance of 7 mm. This is 5% better than a leading registration algorithm while running 14 times faster on whole CT volumes. We demonstrate even greater improvements over detector or similarity-learning alternatives. DLT also generalizes well on an external clinical test set of 100 longitudinal studies, achieving 88% accuracy. Finally, we plug DLT into an automatic tumor monitoring workflow where it leads to an accuracy of 85% in assessing lesion treatment responses, which is only 0.46% lower than the accuracy of manual inputs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge