Lingyun Huang
CVPT: Cross-Attention help Visual Prompt Tuning adapt visual task
Aug 27, 2024Abstract:In recent years, the rapid expansion of model sizes has led to large-scale pre-trained models demonstrating remarkable capabilities. Consequently, there has been a trend towards increasing the scale of models. However, this trend introduces significant challenges, including substantial computational costs of training and transfer to downstream tasks. To address these issues, Parameter-Efficient Fine-Tuning (PEFT) methods have been introduced. These methods optimize large-scale pre-trained models for specific tasks by fine-tuning a select group of parameters. Among these PEFT methods, adapter-based and prompt-based methods are the primary techniques. Specifically, in the field of visual fine-tuning, adapters gain prominence over prompts because of the latter's relatively weaker performance and efficiency. Under the circumstances, we refine the widely-used Visual Prompt Tuning (VPT) method, proposing Cross Visual Prompt Tuning (CVPT). CVPT calculates cross-attention between the prompt tokens and the embedded tokens, which allows us to compute the semantic relationship between them and conduct the fine-tuning of models exactly to adapt visual tasks better. Furthermore, we introduce the weight-sharing mechanism to initialize the parameters of cross-attention, which avoids massive learnable parameters from cross-attention and enhances the representative capability of cross-attention. We conduct comprehensive testing across 25 datasets and the result indicates that CVPT significantly improves VPT's performance and efficiency in visual tasks. For example, on the VTAB-1K benchmark, CVPT outperforms VPT over 4% in average accuracy, rivaling the advanced adapter-based methods in performance and efficiency. Our experiments confirm that prompt-based methods can achieve exceptional results in visual fine-tuning.
Aggregation of Disentanglement: Reconsidering Domain Variations in Domain Generalization
Feb 05, 2023Abstract:Domain Generalization (DG) is a fundamental challenge for machine learning models, which aims to improve model generalization on various domains. Previous methods focus on generating domain invariant features from various source domains. However, we argue that the domain variantions also contain useful information, ie, classification-aware information, for downstream tasks, which has been largely ignored. Different from learning domain invariant features from source domains, we decouple the input images into Domain Expert Features and noise. The proposed domain expert features lie in a learned latent space where the images in each domain can be classified independently, enabling the implicit use of classification-aware domain variations. Based on the analysis, we proposed a novel paradigm called Domain Disentanglement Network (DDN) to disentangle the domain expert features from the source domain images and aggregate the source domain expert features for representing the target test domain. We also propound a new contrastive learning method to guide the domain expert features to form a more balanced and separable feature space. Experiments on the widely-used benchmarks of PACS, VLCS, OfficeHome, DomainNet, and TerraIncognita demonstrate the competitive performance of our method compared to the recently proposed alternatives.
A deep local attention network for pre-operative lymph node metastasis prediction in pancreatic cancer via multiphase CT imaging
Jan 04, 2023



Abstract:Lymph node (LN) metastasis status is one of the most critical prognostic and cancer staging factors for patients with resectable pancreatic ductal adenocarcinoma (PDAC), or in general, for any types of solid malignant tumors. Preoperative prediction of LN metastasis from non-invasive CT imaging is highly desired, as it might be straightforwardly used to guide the following neoadjuvant treatment decision and surgical planning. Most studies only capture the tumor characteristics in CT imaging to implicitly infer LN metastasis and very few work exploit direct LN's CT imaging information. To the best of our knowledge, this is the first work to propose a fully-automated LN segmentation and identification network to directly facilitate the LN metastasis status prediction task. Nevertheless LN segmentation/detection is very challenging since LN can be easily confused with other hard negative anatomic structures (e.g., vessels) from radiological images. We explore the anatomical spatial context priors of pancreatic LN locations by generating a guiding attention map from related organs and vessels to assist segmentation and infer LN status. As such, LN segmentation is impelled to focus on regions that are anatomically adjacent or plausible with respect to the specific organs and vessels. The metastasized LN identification network is trained to classify the segmented LN instances into positives or negatives by reusing the segmentation network as a pre-trained backbone and padding a new classification head. More importantly, we develop a LN metastasis status prediction network that combines the patient-wise aggregation results of LN segmentation/identification and deep imaging features extracted from the tumor region. Extensive quantitative nested five-fold cross-validation is conducted on a discovery dataset of 749 patients with PDAC.
Rethinking Alignment and Uniformity in Unsupervised Image Semantic Segmentation
Nov 26, 2022Abstract:Unsupervised image semantic segmentation(UISS) aims to match low-level visual features with semantic-level representations without outer supervision. In this paper, we address the critical properties from the view of feature alignments and feature uniformity for UISS models. We also make a comparison between UISS and image-wise representation learning. Based on the analysis, we argue that the existing MI-based methods in UISS suffer from representation collapse. By this, we proposed a robust network called Semantic Attention Network(SAN), in which a new module Semantic Attention(SEAT) is proposed to generate pixel-wise and semantic features dynamically. Experimental results on multiple semantic segmentation benchmarks show that our unsupervised segmentation framework specializes in catching semantic representations, which outperforms all the unpretrained and even several pretrained methods.
Comprehensive and Clinically Accurate Head and Neck Organs at Risk Delineation via Stratified Deep Learning: A Large-scale Multi-Institutional Study
Nov 01, 2021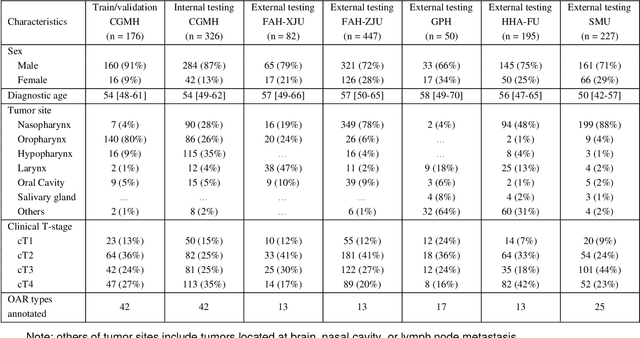
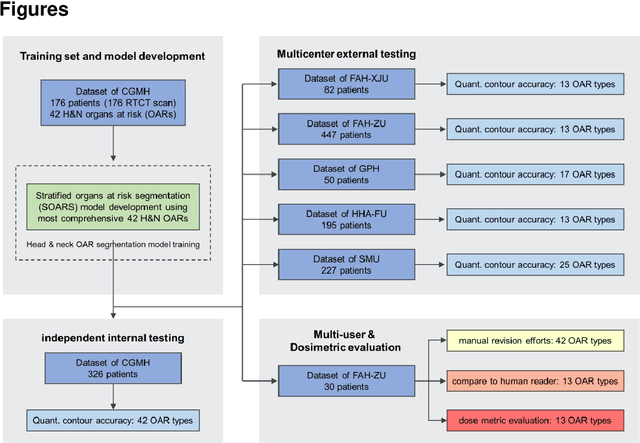
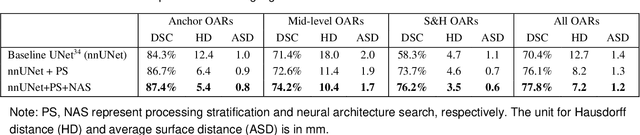
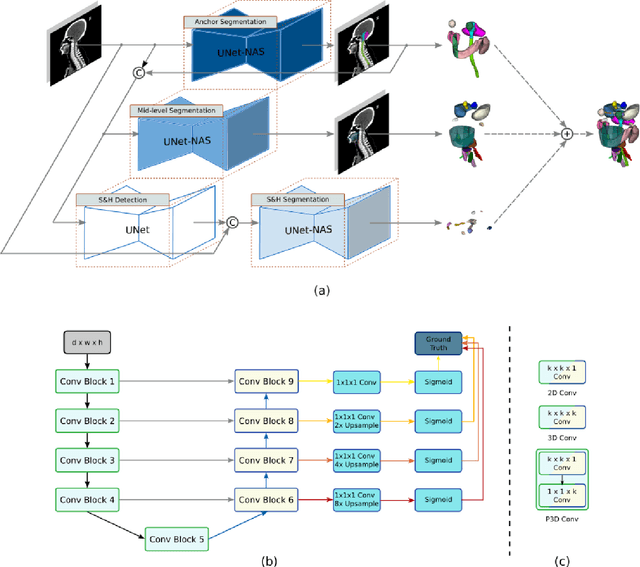
Abstract:Accurate organ at risk (OAR) segmentation is critical to reduce the radiotherapy post-treatment complications. Consensus guidelines recommend a set of more than 40 OARs in the head and neck (H&N) region, however, due to the predictable prohibitive labor-cost of this task, most institutions choose a substantially simplified protocol by delineating a smaller subset of OARs and neglecting the dose distributions associated with other OARs. In this work we propose a novel, automated and highly effective stratified OAR segmentation (SOARS) system using deep learning to precisely delineate a comprehensive set of 42 H&N OARs. SOARS stratifies 42 OARs into anchor, mid-level, and small & hard subcategories, with specifically derived neural network architectures for each category by neural architecture search (NAS) principles. We built SOARS models using 176 training patients in an internal institution and independently evaluated on 1327 external patients across six different institutions. It consistently outperformed other state-of-the-art methods by at least 3-5% in Dice score for each institutional evaluation (up to 36% relative error reduction in other metrics). More importantly, extensive multi-user studies evidently demonstrated that 98% of the SOARS predictions need only very minor or no revisions for direct clinical acceptance (saving 90% radiation oncologists workload), and their segmentation and dosimetric accuracy are within or smaller than the inter-user variation. These findings confirmed the strong clinical applicability of SOARS for the OAR delineation process in H&N cancer radiotherapy workflows, with improved efficiency, comprehensiveness, and quality.
A deep learning pipeline for localization, differentiation, and uncertainty estimation of liver lesions using multi-phasic and multi-sequence MRI
Oct 17, 2021



Abstract:Objectives: to propose a fully-automatic computer-aided diagnosis (CAD) solution for liver lesion characterization, with uncertainty estimation. Methods: we enrolled 400 patients who had either liver resection or a biopsy and was diagnosed with either hepatocellular carcinoma (HCC), intrahepatic cholangiocarcinoma, or secondary metastasis, from 2006 to 2019. Each patient was scanned with T1WI, T2WI, T1WI venous phase (T2WI-V), T1WI arterial phase (T1WI-A), and DWI MRI sequences. We propose a fully-automatic deep CAD pipeline that localizes lesions from 3D MRI studies using key-slice parsing and provides a confidence measure for its diagnoses. We evaluate using five-fold cross validation and compare performance against three radiologists, including a senior hepatology radiologist, a junior hepatology radiologist and an abdominal radiologist. Results: the proposed CAD solution achieves a mean F1 score of 0.62, outperforming the abdominal radiologist (0.47), matching the junior hepatology radiologist (0.61), and underperforming the senior hepatology radiologist (0.68). The CAD system can informatively assess its diagnostic confidence, i.e., when only evaluating on the 70% most confident cases the mean f1 score and sensitivity at 80% specificity for HCC vs. others are boosted from 0.62 to 0.71 and 0.84 to 0.92, respectively. Conclusion: the proposed fully-automatic CAD solution can provide good diagnostic performance with informative confidence assessments in finding and discriminating liver lesions from MRI studies.
Multi-institutional Validation of Two-Streamed Deep Learning Method for Automated Delineation of Esophageal Gross Tumor Volume using planning-CT and FDG-PETCT
Oct 11, 2021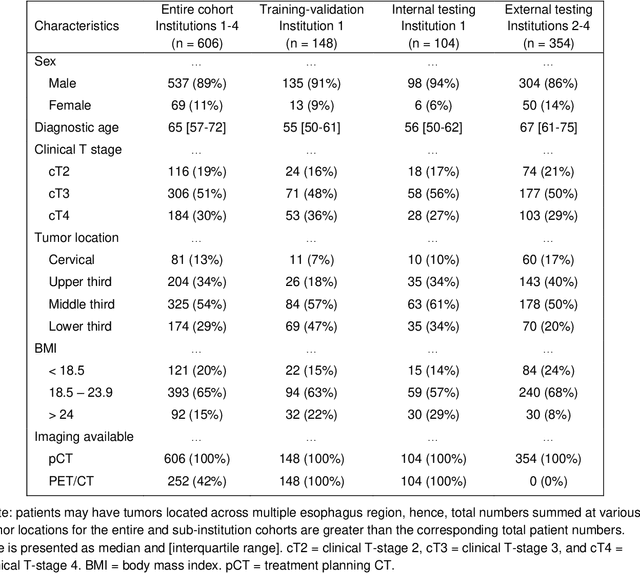
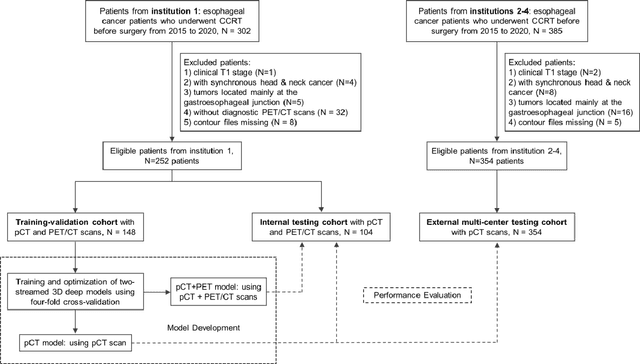
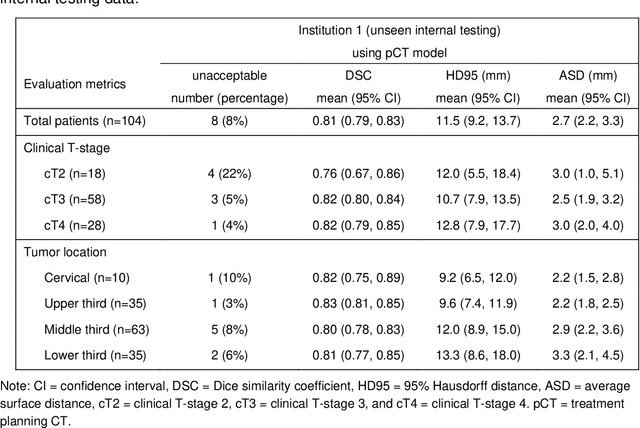
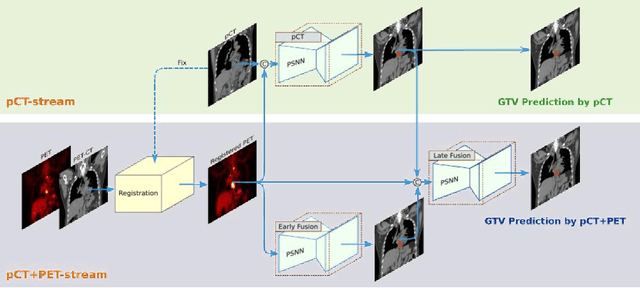
Abstract:Background: The current clinical workflow for esophageal gross tumor volume (GTV) contouring relies on manual delineation of high labor-costs and interuser variability. Purpose: To validate the clinical applicability of a deep learning (DL) multi-modality esophageal GTV contouring model, developed at 1 institution whereas tested at multiple ones. Methods and Materials: We collected 606 esophageal cancer patients from four institutions. 252 institution-1 patients had a treatment planning-CT (pCT) and a pair of diagnostic FDG-PETCT; 354 patients from other 3 institutions had only pCT. A two-streamed DL model for GTV segmentation was developed using pCT and PETCT scans of a 148 patient institution-1 subset. This built model had the flexibility of segmenting GTVs via only pCT or pCT+PETCT combined. For independent evaluation, the rest 104 institution-1 patients behaved as unseen internal testing, and 354 institutions 2-4 patients were used for external testing. We evaluated manual revision degrees by human experts to assess the contour-editing effort. The performance of the deep model was compared against 4 radiation oncologists in a multiuser study with 20 random external patients. Contouring accuracy and time were recorded for the pre-and post-DL assisted delineation process. Results: Our model achieved high segmentation accuracy in internal testing (mean Dice score: 0.81 using pCT and 0.83 using pCT+PET) and generalized well to external evaluation (mean DSC: 0.80). Expert assessment showed that the predicted contours of 88% patients need only minor or no revision. In multi-user evaluation, with the assistance of a deep model, inter-observer variation and required contouring time were reduced by 37.6% and 48.0%, respectively. Conclusions: Deep learning predicted GTV contours were in close agreement with the ground truth and could be adopted clinically with mostly minor or no changes.
SAME: Deformable Image Registration based on Self-supervised Anatomical Embeddings
Sep 23, 2021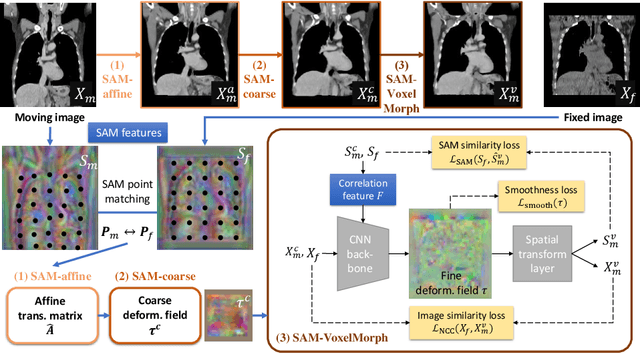
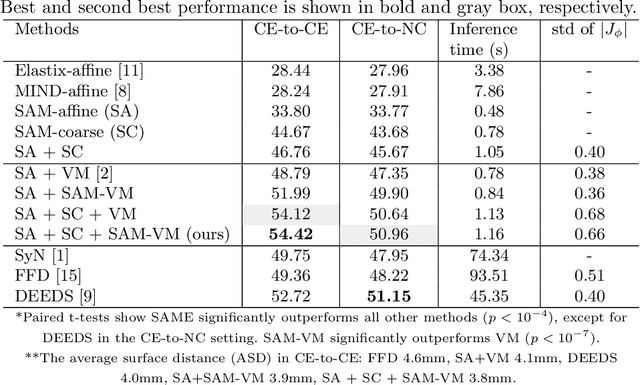
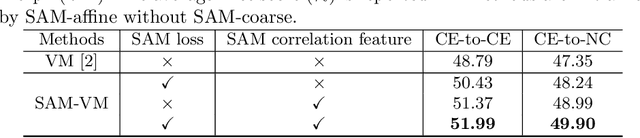

Abstract:In this work, we introduce a fast and accurate method for unsupervised 3D medical image registration. This work is built on top of a recent algorithm SAM, which is capable of computing dense anatomical/semantic correspondences between two images at the pixel level. Our method is named SAME, which breaks down image registration into three steps: affine transformation, coarse deformation, and deep deformable registration. Using SAM embeddings, we enhance these steps by finding more coherent correspondences, and providing features and a loss function with better semantic guidance. We collect a multi-phase chest computed tomography dataset with 35 annotated organs for each patient and conduct inter-subject registration for quantitative evaluation. Results show that SAME outperforms widely-used traditional registration techniques (Elastix FFD, ANTs SyN) and learning based VoxelMorph method by at least 4.7% and 2.7% in Dice scores for two separate tasks of within-contrast-phase and across-contrast-phase registration, respectively. SAME achieves the comparable performance to the best traditional registration method, DEEDS (from our evaluation), while being orders of magnitude faster (from 45 seconds to 1.2 seconds).
DeepStationing: Thoracic Lymph Node Station Parsing in CT Scans using Anatomical Context Encoding and Key Organ Auto-Search
Sep 20, 2021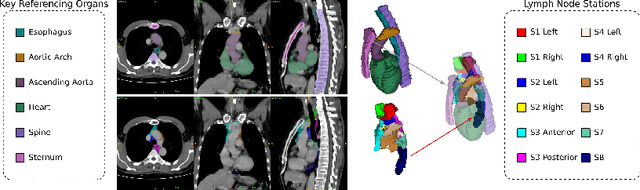
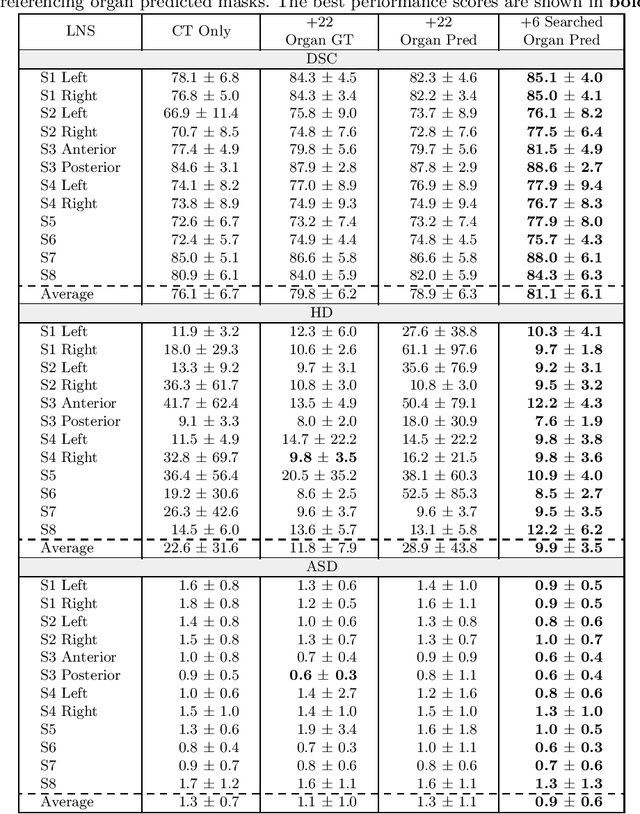
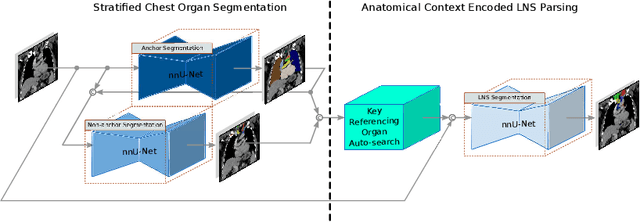

Abstract:Lymph node station (LNS) delineation from computed tomography (CT) scans is an indispensable step in radiation oncology workflow. High inter-user variabilities across oncologists and prohibitive laboring costs motivated the automated approach. Previous works exploit anatomical priors to infer LNS based on predefined ad-hoc margins. However, without voxel-level supervision, the performance is severely limited. LNS is highly context-dependent - LNS boundaries are constrained by anatomical organs - we formulate it as a deep spatial and contextual parsing problem via encoded anatomical organs. This permits the deep network to better learn from both CT appearance and organ context. We develop a stratified referencing organ segmentation protocol that divides the organs into anchor and non-anchor categories and uses the former's predictions to guide the later segmentation. We further develop an auto-search module to identify the key organs that opt for the optimal LNS parsing performance. Extensive four-fold cross-validation experiments on a dataset of 98 esophageal cancer patients (with the most comprehensive set of 12 LNSs + 22 organs in thoracic region to date) are conducted. Our LNS parsing model produces significant performance improvements, with an average Dice score of 81.1% +/- 6.1%, which is 5.0% and 19.2% higher over the pure CT-based deep model and the previous representative approach, respectively.
Lesion Segmentation and RECIST Diameter Prediction via Click-driven Attention and Dual-path Connection
May 05, 2021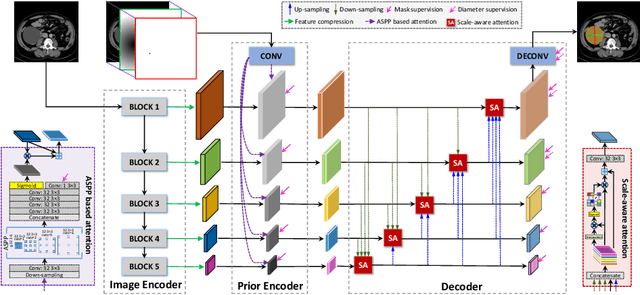
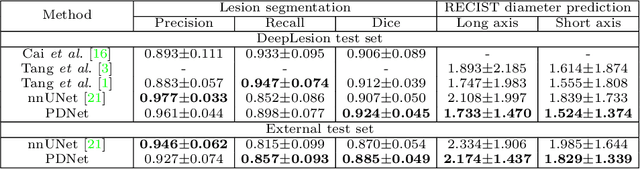
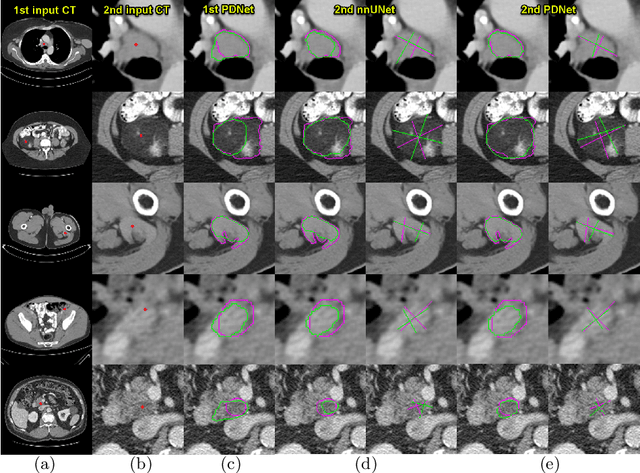
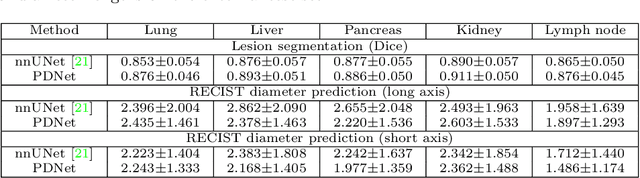
Abstract:Measuring lesion size is an important step to assess tumor growth and monitor disease progression and therapy response in oncology image analysis. Although it is tedious and highly time-consuming, radiologists have to work on this task by using RECIST criteria (Response Evaluation Criteria In Solid Tumors) routinely and manually. Even though lesion segmentation may be the more accurate and clinically more valuable means, physicians can not manually segment lesions as now since much more heavy laboring will be required. In this paper, we present a prior-guided dual-path network (PDNet) to segment common types of lesions throughout the whole body and predict their RECIST diameters accurately and automatically. Similar to [1], a click guidance from radiologists is the only requirement. There are two key characteristics in PDNet: 1) Learning lesion-specific attention matrices in parallel from the click prior information by the proposed prior encoder, named click-driven attention; 2) Aggregating the extracted multi-scale features comprehensively by introducing top-down and bottom-up connections in the proposed decoder, named dual-path connection. Experiments show the superiority of our proposed PDNet in lesion segmentation and RECIST diameter prediction using the DeepLesion dataset and an external test set. PDNet learns comprehensive and representative deep image features for our tasks and produces more accurate results on both lesion segmentation and RECIST diameter prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge