Ling Peng
Comprehensive and Clinically Accurate Head and Neck Organs at Risk Delineation via Stratified Deep Learning: A Large-scale Multi-Institutional Study
Nov 01, 2021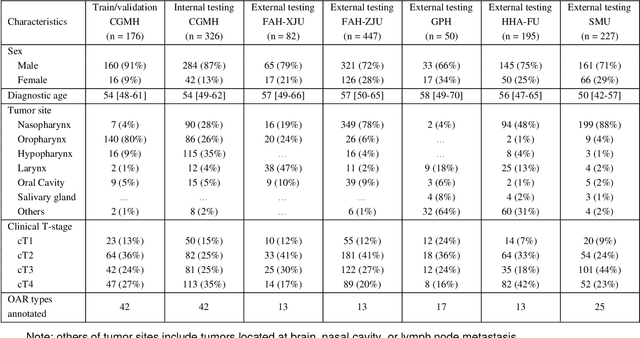
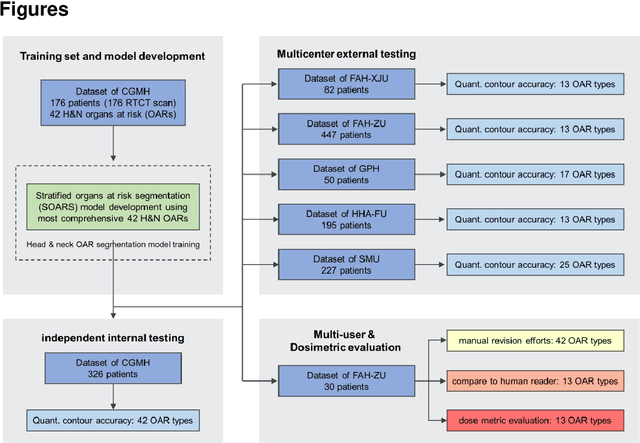
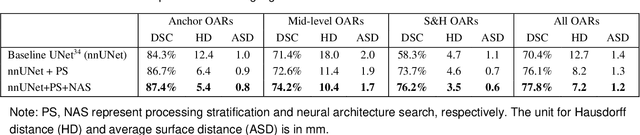
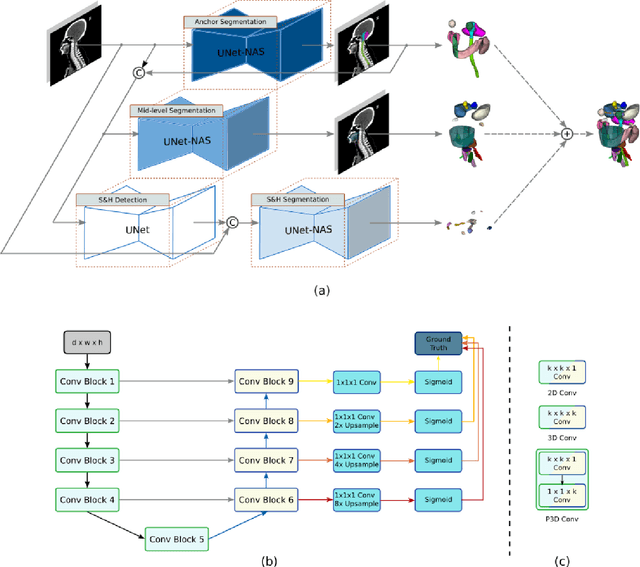
Abstract:Accurate organ at risk (OAR) segmentation is critical to reduce the radiotherapy post-treatment complications. Consensus guidelines recommend a set of more than 40 OARs in the head and neck (H&N) region, however, due to the predictable prohibitive labor-cost of this task, most institutions choose a substantially simplified protocol by delineating a smaller subset of OARs and neglecting the dose distributions associated with other OARs. In this work we propose a novel, automated and highly effective stratified OAR segmentation (SOARS) system using deep learning to precisely delineate a comprehensive set of 42 H&N OARs. SOARS stratifies 42 OARs into anchor, mid-level, and small & hard subcategories, with specifically derived neural network architectures for each category by neural architecture search (NAS) principles. We built SOARS models using 176 training patients in an internal institution and independently evaluated on 1327 external patients across six different institutions. It consistently outperformed other state-of-the-art methods by at least 3-5% in Dice score for each institutional evaluation (up to 36% relative error reduction in other metrics). More importantly, extensive multi-user studies evidently demonstrated that 98% of the SOARS predictions need only very minor or no revisions for direct clinical acceptance (saving 90% radiation oncologists workload), and their segmentation and dosimetric accuracy are within or smaller than the inter-user variation. These findings confirmed the strong clinical applicability of SOARS for the OAR delineation process in H&N cancer radiotherapy workflows, with improved efficiency, comprehensiveness, and quality.
Multi-institutional Validation of Two-Streamed Deep Learning Method for Automated Delineation of Esophageal Gross Tumor Volume using planning-CT and FDG-PETCT
Oct 11, 2021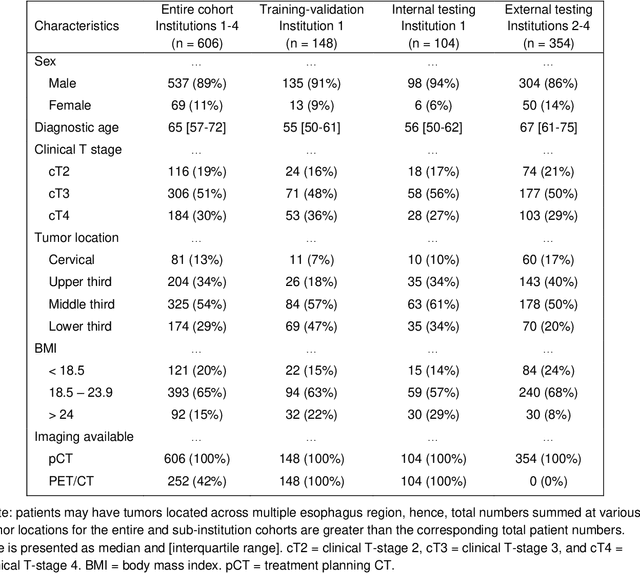
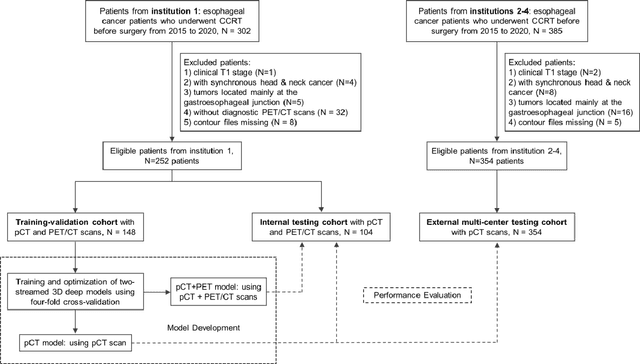
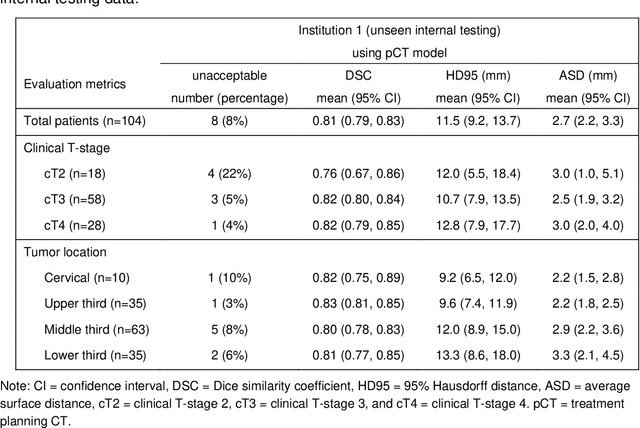
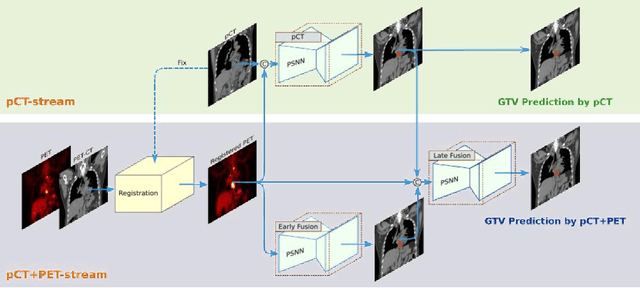
Abstract:Background: The current clinical workflow for esophageal gross tumor volume (GTV) contouring relies on manual delineation of high labor-costs and interuser variability. Purpose: To validate the clinical applicability of a deep learning (DL) multi-modality esophageal GTV contouring model, developed at 1 institution whereas tested at multiple ones. Methods and Materials: We collected 606 esophageal cancer patients from four institutions. 252 institution-1 patients had a treatment planning-CT (pCT) and a pair of diagnostic FDG-PETCT; 354 patients from other 3 institutions had only pCT. A two-streamed DL model for GTV segmentation was developed using pCT and PETCT scans of a 148 patient institution-1 subset. This built model had the flexibility of segmenting GTVs via only pCT or pCT+PETCT combined. For independent evaluation, the rest 104 institution-1 patients behaved as unseen internal testing, and 354 institutions 2-4 patients were used for external testing. We evaluated manual revision degrees by human experts to assess the contour-editing effort. The performance of the deep model was compared against 4 radiation oncologists in a multiuser study with 20 random external patients. Contouring accuracy and time were recorded for the pre-and post-DL assisted delineation process. Results: Our model achieved high segmentation accuracy in internal testing (mean Dice score: 0.81 using pCT and 0.83 using pCT+PET) and generalized well to external evaluation (mean DSC: 0.80). Expert assessment showed that the predicted contours of 88% patients need only minor or no revision. In multi-user evaluation, with the assistance of a deep model, inter-observer variation and required contouring time were reduced by 37.6% and 48.0%, respectively. Conclusions: Deep learning predicted GTV contours were in close agreement with the ground truth and could be adopted clinically with mostly minor or no changes.
DeepStationing: Thoracic Lymph Node Station Parsing in CT Scans using Anatomical Context Encoding and Key Organ Auto-Search
Sep 20, 2021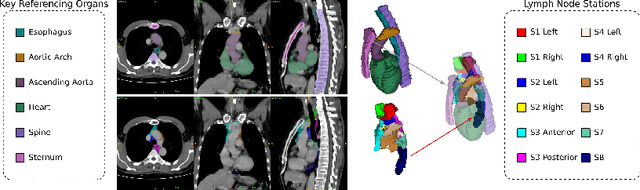
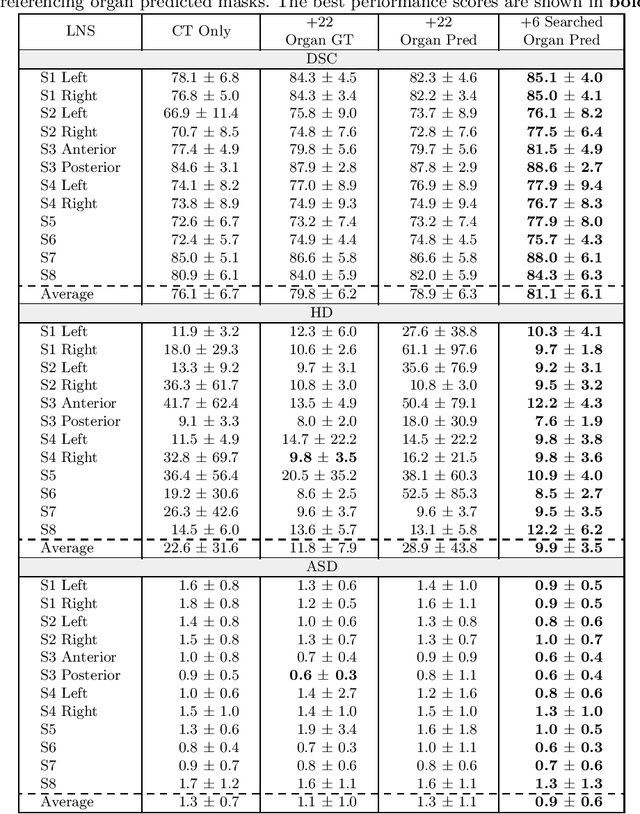
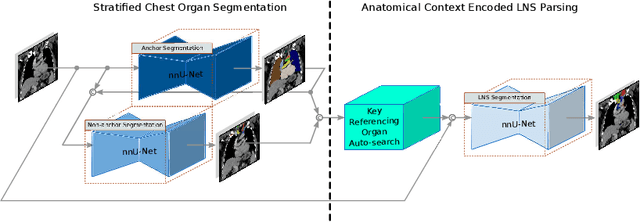

Abstract:Lymph node station (LNS) delineation from computed tomography (CT) scans is an indispensable step in radiation oncology workflow. High inter-user variabilities across oncologists and prohibitive laboring costs motivated the automated approach. Previous works exploit anatomical priors to infer LNS based on predefined ad-hoc margins. However, without voxel-level supervision, the performance is severely limited. LNS is highly context-dependent - LNS boundaries are constrained by anatomical organs - we formulate it as a deep spatial and contextual parsing problem via encoded anatomical organs. This permits the deep network to better learn from both CT appearance and organ context. We develop a stratified referencing organ segmentation protocol that divides the organs into anchor and non-anchor categories and uses the former's predictions to guide the later segmentation. We further develop an auto-search module to identify the key organs that opt for the optimal LNS parsing performance. Extensive four-fold cross-validation experiments on a dataset of 98 esophageal cancer patients (with the most comprehensive set of 12 LNSs + 22 organs in thoracic region to date) are conducted. Our LNS parsing model produces significant performance improvements, with an average Dice score of 81.1% +/- 6.1%, which is 5.0% and 19.2% higher over the pure CT-based deep model and the previous representative approach, respectively.
Airborne LiDAR Point Cloud Classification with Graph Attention Convolution Neural Network
Apr 20, 2020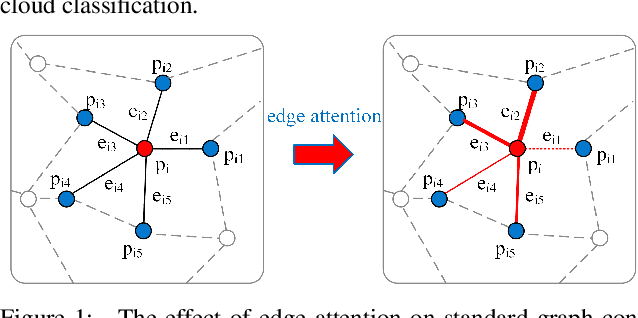
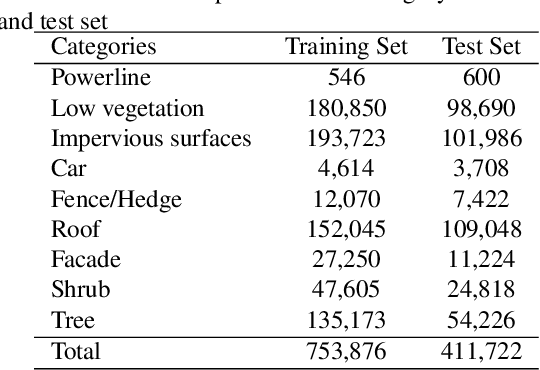
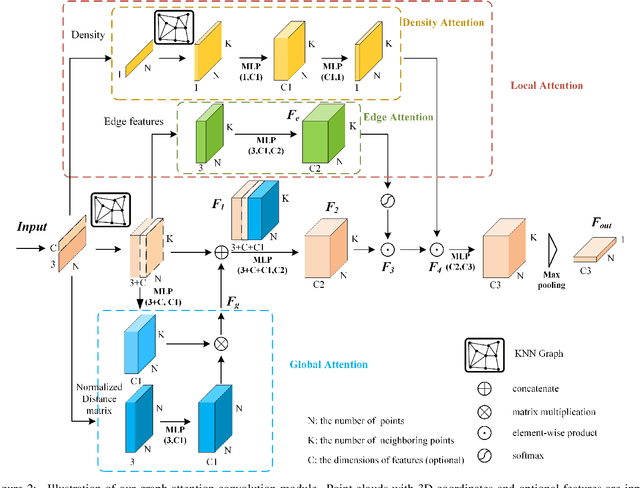
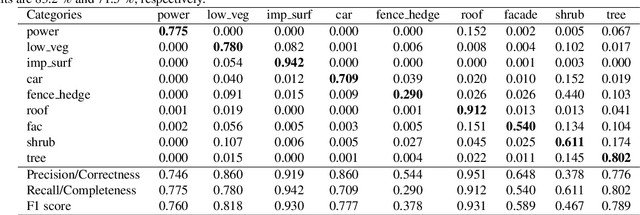
Abstract:Airborne light detection and ranging (LiDAR) plays an increasingly significant role in urban planning, topographic mapping, environmental monitoring, power line detection and other fields thanks to its capability to quickly acquire large-scale and high-precision ground information. To achieve point cloud classification, previous studies proposed point cloud deep learning models that can directly process raw point clouds based on PointNet-like architectures. And some recent works proposed graph convolution neural network based on the inherent topology of point clouds. However, the above point cloud deep learning models only pay attention to exploring local geometric structures, yet ignore global contextual relationships among all points. In this paper, we present a graph attention convolution neural network (GACNN) that can be directly applied to the classification of unstructured 3D point clouds obtained by airborne LiDAR. Specifically, we first introduce a graph attention convolution module that incorporates global contextual information and local structural features. Based on the proposed graph attention convolution module, we further design an end-to-end encoder-decoder network, named GACNN, to capture multiscale features of the point clouds and therefore enable more accurate airborne point cloud classification. Experiments on the ISPRS 3D labeling dataset show that the proposed model achieves a new state-of-the-art performance in terms of average F1 score (71.5\%) and a satisfying overall accuracy (83.2\%). Additionally, experiments further conducted on the 2019 Data Fusion Contest Dataset by comparing with other prevalent point cloud deep learning models demonstrate the favorable generalization capability of the proposed model.
Directionally Constrained Fully Convolutional Neural Network For Airborne Lidar Point Cloud Classification
Aug 19, 2019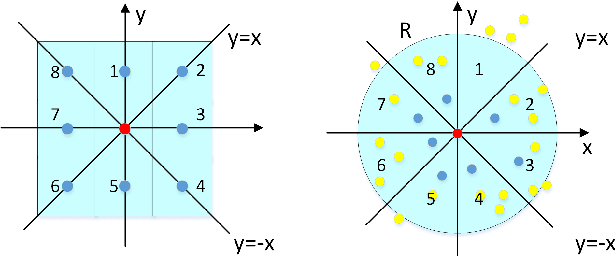
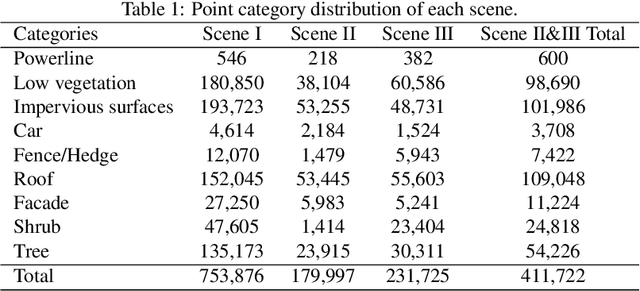
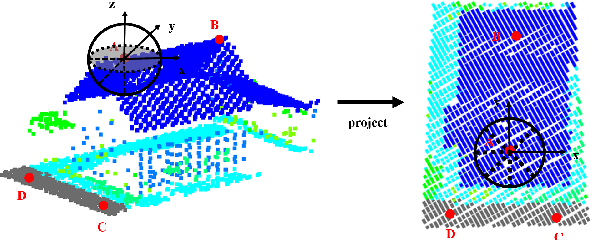
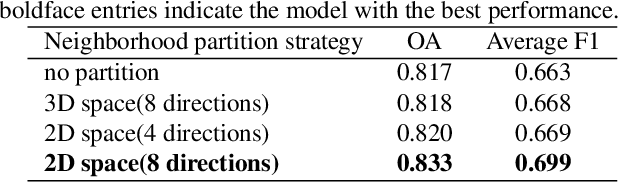
Abstract:Point cloud classification plays an important role in a wide range of airborne light detection and ranging (LiDAR) applications, such as topographic mapping, forest monitoring, power line detection, and road detection. However, due to the sensor noise, high redundancy, incompleteness, and complexity of airborne LiDAR systems, point cloud classification is challenging. In this paper, we proposed a directionally constrained fully convolutional neural network (D-FCN) that can take the original 3D coordinates and LiDAR intensity as input; thus, it can directly apply to unstructured 3D point clouds for semantic labeling. Specifically, we first introduce a novel directionally constrained point convolution (D-Conv) module to extract locally representative features of 3D point sets from the projected 2D receptive fields. To make full use of the orientation information of neighborhood points, the proposed D-Conv module performs convolution in an orientation-aware manner by using a directionally constrained nearest neighborhood search. Then, we designed a multiscale fully convolutional neural network with downsampling and upsampling blocks to enable multiscale point feature learning. The proposed D-FCN model can therefore process input point cloud with arbitrary sizes and directly predict the semantic labels for all the input points in an end-to-end manner. Without involving additional geometry features as input, the proposed method has demonstrated superior performance on the International Society for Photogrammetry and Remote Sensing (ISPRS) 3D labeling benchmark dataset. The results show that our model has achieved a new state-of-the-art level of performance with an average F1 score of 70.7%, and it has improved the performance by a large margin on categories with a small number of points (such as powerline, car, and facade).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge