Yanping Ren
Comprehensive and Clinically Accurate Head and Neck Organs at Risk Delineation via Stratified Deep Learning: A Large-scale Multi-Institutional Study
Nov 01, 2021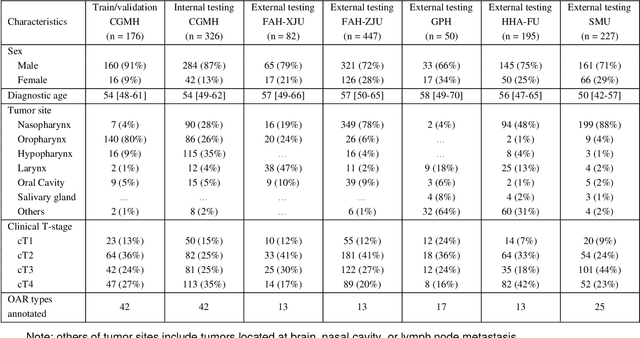
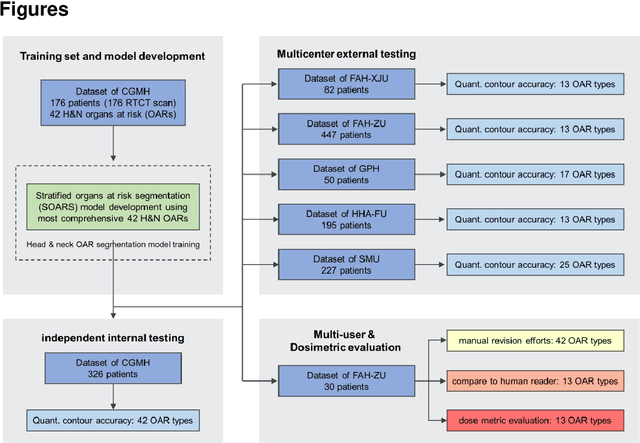
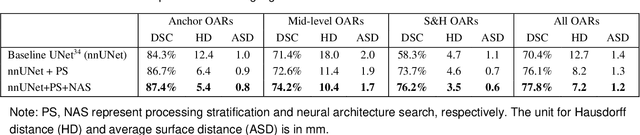
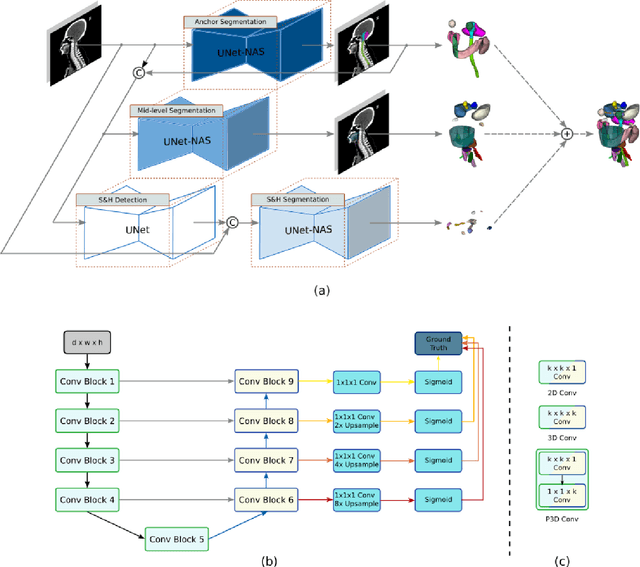
Abstract:Accurate organ at risk (OAR) segmentation is critical to reduce the radiotherapy post-treatment complications. Consensus guidelines recommend a set of more than 40 OARs in the head and neck (H&N) region, however, due to the predictable prohibitive labor-cost of this task, most institutions choose a substantially simplified protocol by delineating a smaller subset of OARs and neglecting the dose distributions associated with other OARs. In this work we propose a novel, automated and highly effective stratified OAR segmentation (SOARS) system using deep learning to precisely delineate a comprehensive set of 42 H&N OARs. SOARS stratifies 42 OARs into anchor, mid-level, and small & hard subcategories, with specifically derived neural network architectures for each category by neural architecture search (NAS) principles. We built SOARS models using 176 training patients in an internal institution and independently evaluated on 1327 external patients across six different institutions. It consistently outperformed other state-of-the-art methods by at least 3-5% in Dice score for each institutional evaluation (up to 36% relative error reduction in other metrics). More importantly, extensive multi-user studies evidently demonstrated that 98% of the SOARS predictions need only very minor or no revisions for direct clinical acceptance (saving 90% radiation oncologists workload), and their segmentation and dosimetric accuracy are within or smaller than the inter-user variation. These findings confirmed the strong clinical applicability of SOARS for the OAR delineation process in H&N cancer radiotherapy workflows, with improved efficiency, comprehensiveness, and quality.
Multi-institutional Validation of Two-Streamed Deep Learning Method for Automated Delineation of Esophageal Gross Tumor Volume using planning-CT and FDG-PETCT
Oct 11, 2021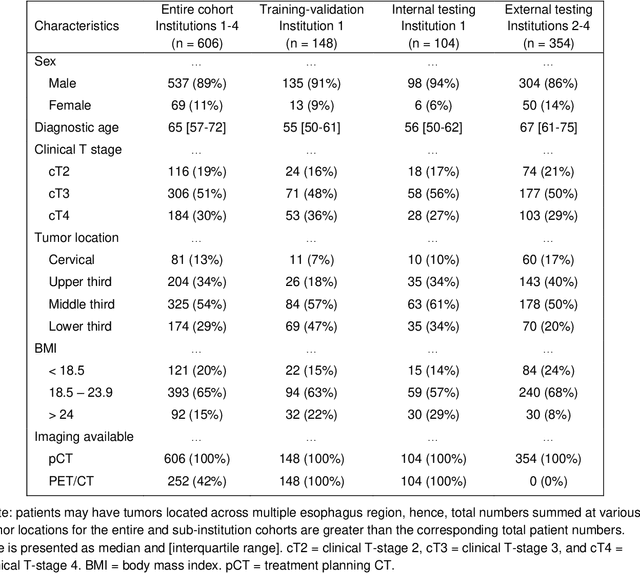
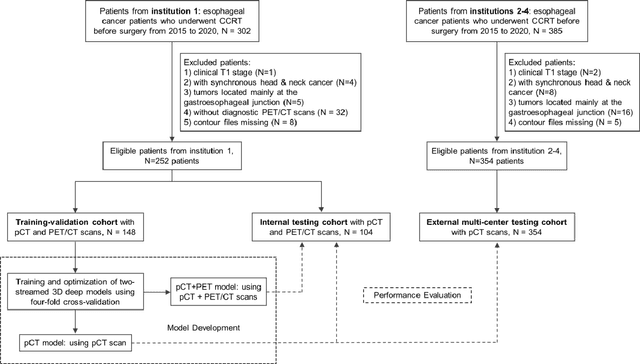
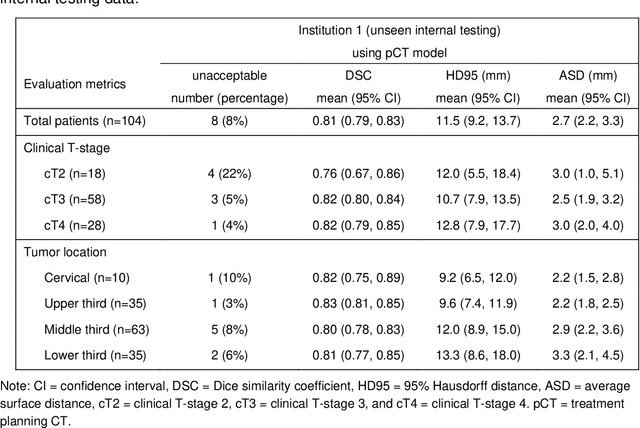
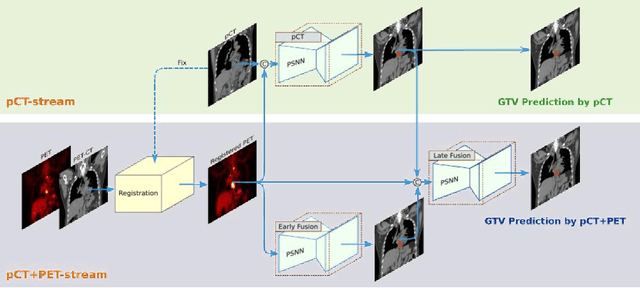
Abstract:Background: The current clinical workflow for esophageal gross tumor volume (GTV) contouring relies on manual delineation of high labor-costs and interuser variability. Purpose: To validate the clinical applicability of a deep learning (DL) multi-modality esophageal GTV contouring model, developed at 1 institution whereas tested at multiple ones. Methods and Materials: We collected 606 esophageal cancer patients from four institutions. 252 institution-1 patients had a treatment planning-CT (pCT) and a pair of diagnostic FDG-PETCT; 354 patients from other 3 institutions had only pCT. A two-streamed DL model for GTV segmentation was developed using pCT and PETCT scans of a 148 patient institution-1 subset. This built model had the flexibility of segmenting GTVs via only pCT or pCT+PETCT combined. For independent evaluation, the rest 104 institution-1 patients behaved as unseen internal testing, and 354 institutions 2-4 patients were used for external testing. We evaluated manual revision degrees by human experts to assess the contour-editing effort. The performance of the deep model was compared against 4 radiation oncologists in a multiuser study with 20 random external patients. Contouring accuracy and time were recorded for the pre-and post-DL assisted delineation process. Results: Our model achieved high segmentation accuracy in internal testing (mean Dice score: 0.81 using pCT and 0.83 using pCT+PET) and generalized well to external evaluation (mean DSC: 0.80). Expert assessment showed that the predicted contours of 88% patients need only minor or no revision. In multi-user evaluation, with the assistance of a deep model, inter-observer variation and required contouring time were reduced by 37.6% and 48.0%, respectively. Conclusions: Deep learning predicted GTV contours were in close agreement with the ground truth and could be adopted clinically with mostly minor or no changes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge