Ping-Ching Pai
Multi-institutional Validation of Two-Streamed Deep Learning Method for Automated Delineation of Esophageal Gross Tumor Volume using planning-CT and FDG-PETCT
Oct 11, 2021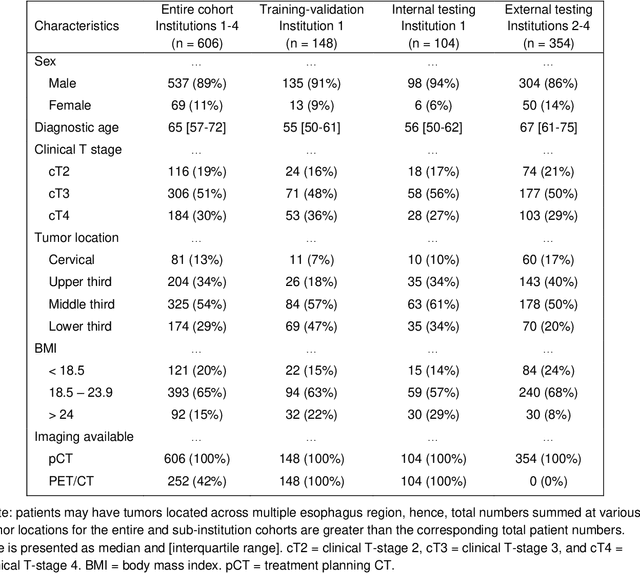
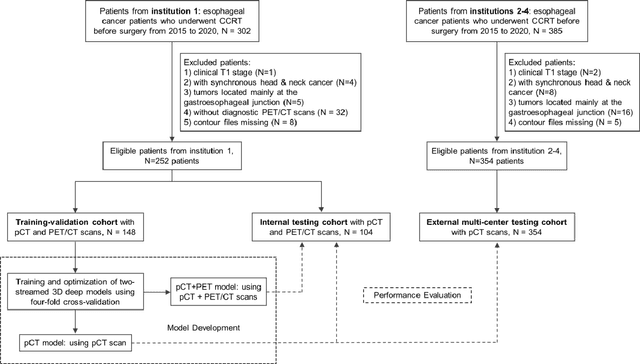
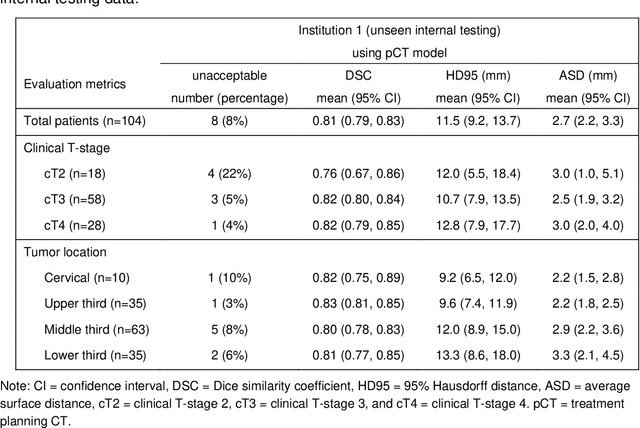
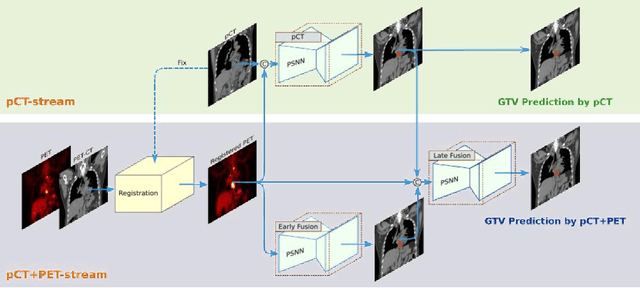
Abstract:Background: The current clinical workflow for esophageal gross tumor volume (GTV) contouring relies on manual delineation of high labor-costs and interuser variability. Purpose: To validate the clinical applicability of a deep learning (DL) multi-modality esophageal GTV contouring model, developed at 1 institution whereas tested at multiple ones. Methods and Materials: We collected 606 esophageal cancer patients from four institutions. 252 institution-1 patients had a treatment planning-CT (pCT) and a pair of diagnostic FDG-PETCT; 354 patients from other 3 institutions had only pCT. A two-streamed DL model for GTV segmentation was developed using pCT and PETCT scans of a 148 patient institution-1 subset. This built model had the flexibility of segmenting GTVs via only pCT or pCT+PETCT combined. For independent evaluation, the rest 104 institution-1 patients behaved as unseen internal testing, and 354 institutions 2-4 patients were used for external testing. We evaluated manual revision degrees by human experts to assess the contour-editing effort. The performance of the deep model was compared against 4 radiation oncologists in a multiuser study with 20 random external patients. Contouring accuracy and time were recorded for the pre-and post-DL assisted delineation process. Results: Our model achieved high segmentation accuracy in internal testing (mean Dice score: 0.81 using pCT and 0.83 using pCT+PET) and generalized well to external evaluation (mean DSC: 0.80). Expert assessment showed that the predicted contours of 88% patients need only minor or no revision. In multi-user evaluation, with the assistance of a deep model, inter-observer variation and required contouring time were reduced by 37.6% and 48.0%, respectively. Conclusions: Deep learning predicted GTV contours were in close agreement with the ground truth and could be adopted clinically with mostly minor or no changes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge