Adam P. Harrison
Localized Adversarial Domain Generalization
May 09, 2022



Abstract:Deep learning methods can struggle to handle domain shifts not seen in training data, which can cause them to not generalize well to unseen domains. This has led to research attention on domain generalization (DG), which aims to the model's generalization ability to out-of-distribution. Adversarial domain generalization is a popular approach to DG, but conventional approaches (1) struggle to sufficiently align features so that local neighborhoods are mixed across domains; and (2) can suffer from feature space over collapse which can threaten generalization performance. To address these limitations, we propose localized adversarial domain generalization with space compactness maintenance~(LADG) which constitutes two major contributions. First, we propose an adversarial localized classifier as the domain discriminator, along with a principled primary branch. This constructs a min-max game whereby the aim of the featurizer is to produce locally mixed domains. Second, we propose to use a coding-rate loss to alleviate feature space over collapse. We conduct comprehensive experiments on the Wilds DG benchmark to validate our approach, where LADG outperforms leading competitors on most datasets.
Comprehensive and Clinically Accurate Head and Neck Organs at Risk Delineation via Stratified Deep Learning: A Large-scale Multi-Institutional Study
Nov 01, 2021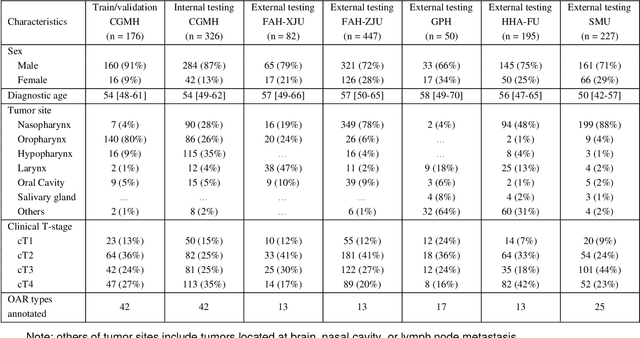
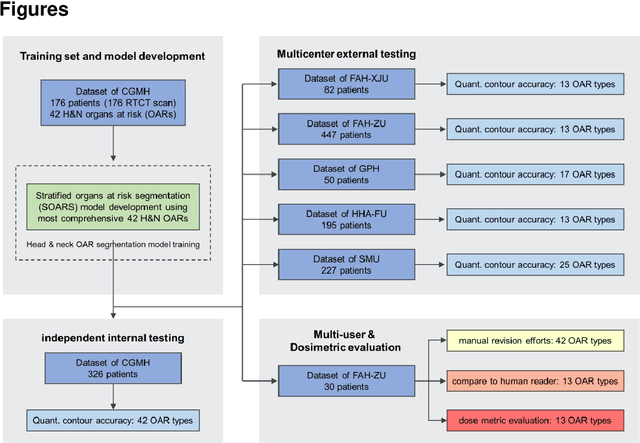
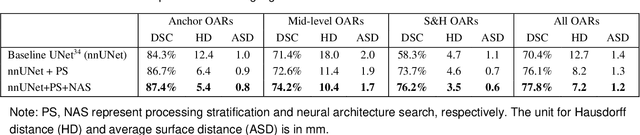
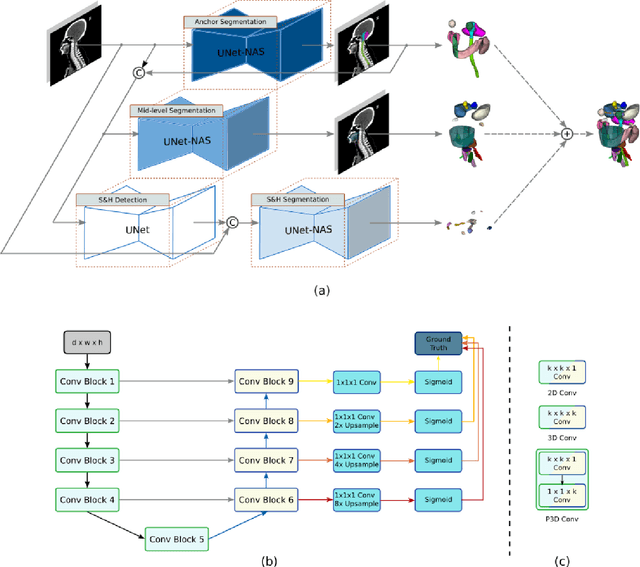
Abstract:Accurate organ at risk (OAR) segmentation is critical to reduce the radiotherapy post-treatment complications. Consensus guidelines recommend a set of more than 40 OARs in the head and neck (H&N) region, however, due to the predictable prohibitive labor-cost of this task, most institutions choose a substantially simplified protocol by delineating a smaller subset of OARs and neglecting the dose distributions associated with other OARs. In this work we propose a novel, automated and highly effective stratified OAR segmentation (SOARS) system using deep learning to precisely delineate a comprehensive set of 42 H&N OARs. SOARS stratifies 42 OARs into anchor, mid-level, and small & hard subcategories, with specifically derived neural network architectures for each category by neural architecture search (NAS) principles. We built SOARS models using 176 training patients in an internal institution and independently evaluated on 1327 external patients across six different institutions. It consistently outperformed other state-of-the-art methods by at least 3-5% in Dice score for each institutional evaluation (up to 36% relative error reduction in other metrics). More importantly, extensive multi-user studies evidently demonstrated that 98% of the SOARS predictions need only very minor or no revisions for direct clinical acceptance (saving 90% radiation oncologists workload), and their segmentation and dosimetric accuracy are within or smaller than the inter-user variation. These findings confirmed the strong clinical applicability of SOARS for the OAR delineation process in H&N cancer radiotherapy workflows, with improved efficiency, comprehensiveness, and quality.
A deep learning pipeline for localization, differentiation, and uncertainty estimation of liver lesions using multi-phasic and multi-sequence MRI
Oct 17, 2021



Abstract:Objectives: to propose a fully-automatic computer-aided diagnosis (CAD) solution for liver lesion characterization, with uncertainty estimation. Methods: we enrolled 400 patients who had either liver resection or a biopsy and was diagnosed with either hepatocellular carcinoma (HCC), intrahepatic cholangiocarcinoma, or secondary metastasis, from 2006 to 2019. Each patient was scanned with T1WI, T2WI, T1WI venous phase (T2WI-V), T1WI arterial phase (T1WI-A), and DWI MRI sequences. We propose a fully-automatic deep CAD pipeline that localizes lesions from 3D MRI studies using key-slice parsing and provides a confidence measure for its diagnoses. We evaluate using five-fold cross validation and compare performance against three radiologists, including a senior hepatology radiologist, a junior hepatology radiologist and an abdominal radiologist. Results: the proposed CAD solution achieves a mean F1 score of 0.62, outperforming the abdominal radiologist (0.47), matching the junior hepatology radiologist (0.61), and underperforming the senior hepatology radiologist (0.68). The CAD system can informatively assess its diagnostic confidence, i.e., when only evaluating on the 70% most confident cases the mean f1 score and sensitivity at 80% specificity for HCC vs. others are boosted from 0.62 to 0.71 and 0.84 to 0.92, respectively. Conclusion: the proposed fully-automatic CAD solution can provide good diagnostic performance with informative confidence assessments in finding and discriminating liver lesions from MRI studies.
Accurate and Generalizable Quantitative Scoring of Liver Steatosis from Ultrasound Images via Scalable Deep Learning
Oct 12, 2021


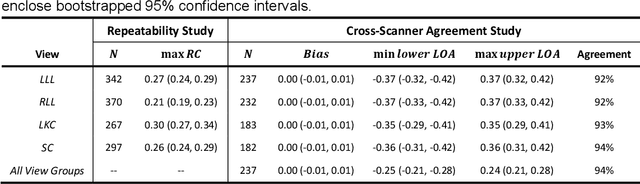
Abstract:Background & Aims: Hepatic steatosis is a major cause of chronic liver disease. 2D ultrasound is the most widely used non-invasive tool for screening and monitoring, but associated diagnoses are highly subjective. We developed a scalable deep learning (DL) algorithm for quantitative scoring of liver steatosis from 2D ultrasound images. Approach & Results: Using retrospectively collected multi-view ultrasound data from 3,310 patients, 19,513 studies, and 228,075 images, we trained a DL algorithm to diagnose steatosis stages (healthy, mild, moderate, or severe) from ultrasound diagnoses. Performance was validated on two multi-scanner unblinded and blinded (initially to DL developer) histology-proven cohorts (147 and 112 patients) with histopathology fatty cell percentage diagnoses, and a subset with FibroScan diagnoses. We also quantified reliability across scanners and viewpoints. Results were evaluated using Bland-Altman and receiver operating characteristic (ROC) analysis. The DL algorithm demonstrates repeatable measurements with a moderate number of images (3 for each viewpoint) and high agreement across 3 premium ultrasound scanners. High diagnostic performance was observed across all viewpoints: area under the curves of the ROC to classify >=mild, >=moderate, =severe steatosis grades were 0.85, 0.90, and 0.93, respectively. The DL algorithm outperformed or performed at least comparably to FibroScan with statistically significant improvements for all levels on the unblinded histology-proven cohort, and for =severe steatosis on the blinded histology-proven cohort. Conclusions: The DL algorithm provides a reliable quantitative steatosis assessment across view and scanners on two multi-scanner cohorts. Diagnostic performance was high with comparable or better performance than FibroScan.
A Flexible Three-Dimensional Hetero-phase Computed Tomography Hepatocellular Carcinoma (HCC) Detection Algorithm for Generalizable and Practical HCC Screening
Aug 17, 2021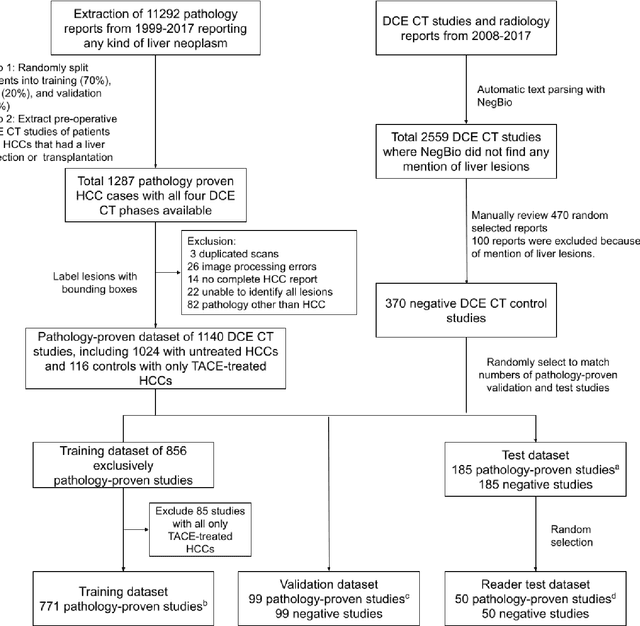
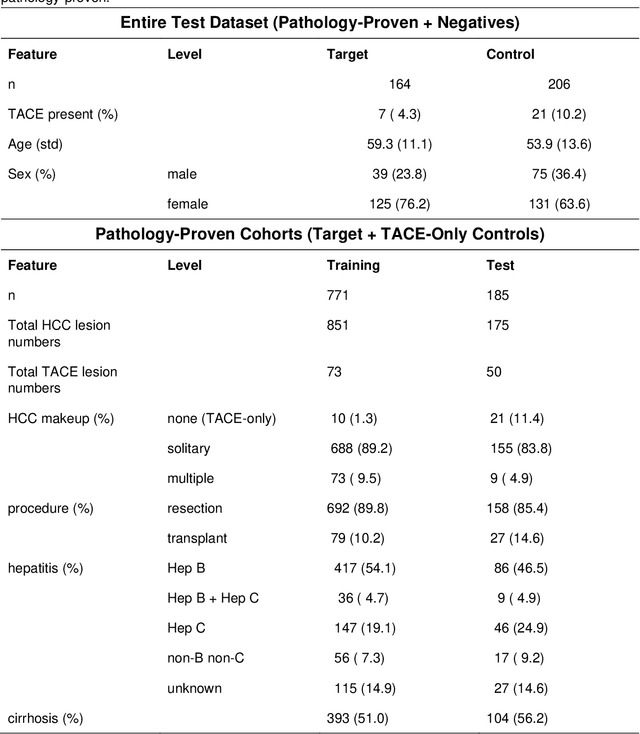
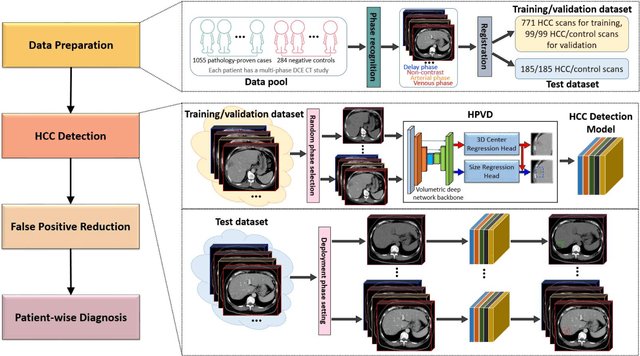
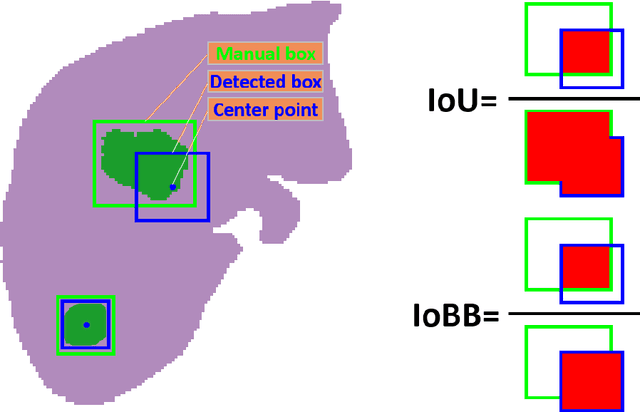
Abstract:Hepatocellular carcinoma (HCC) can be potentially discovered from abdominal computed tomography (CT) studies under varied clinical scenarios, e.g., fully dynamic contrast enhanced (DCE) studies, non-contrast (NC) plus venous phase (VP) abdominal studies, or NC-only studies. We develop a flexible three-dimensional deep algorithm, called hetero-phase volumetric detection (HPVD), that can accept any combination of contrast-phase inputs and with adjustable sensitivity depending on the clinical purpose. We trained HPVD on 771 DCE CT scans to detect HCCs and tested on external 164 positives and 206 controls, respectively. We compare performance against six clinical readers, including two radiologists, two hepato-pancreatico-biliary (HPB) surgeons, and two hepatologists. The area under curve (AUC) of the localization receiver operating characteristic (LROC) for NC-only, NC plus VP, and full DCE CT yielded 0.71, 0.81, 0.89 respectively. At a high sensitivity operating point of 80% on DCE CT, HPVD achieved 97% specificity, which is comparable to measured physician performance. We also demonstrate performance improvements over more typical and less flexible non hetero-phase detectors. Thus, we demonstrate that a single deep learning algorithm can be effectively applied to diverse HCC detection clinical scenarios.
Learning from Subjective Ratings Using Auto-Decoded Deep Latent Embeddings
Apr 16, 2021



Abstract:Depending on the application, radiological diagnoses can be associated with high inter- and intra-rater variabilities. Most computer-aided diagnosis (CAD) solutions treat such data as incontrovertible, exposing learning algorithms to considerable and possibly contradictory label noise and biases. Thus, managing subjectivity in labels is a fundamental problem in medical imaging analysis. To address this challenge, we introduce auto-decoded deep latent embeddings (ADDLE), which explicitly models the tendencies of each rater using an auto-decoder framework. After a simple linear transformation, the latent variables can be injected into any backbone at any and multiple points, allowing the model to account for rater-specific effects on the diagnosis. Importantly, ADDLE does not expect multiple raters per image in training, meaning it can readily learn from data mined from hospital archives. Moreover, the complexity of training ADDLE does not increase as more raters are added. During inference each rater can be simulated and a 'mean' or 'greedy' virtual rating can be produced. We test ADDLE on the problem of liver steatosis diagnosis from 2D ultrasound (US) by collecting 46 084 studies along with clinical US diagnoses originating from 65 different raters. We evaluated diagnostic performance using a separate dataset with gold-standard biopsy diagnoses. ADDLE can improve the partial areas under the curve (AUCs) for diagnosing severe steatosis by 10.5% over standard classifiers while outperforming other annotator-noise approaches, including those requiring 65 times the parameters.
Deep Implicit Statistical Shape Models for 3D Medical Image Delineation
Apr 07, 2021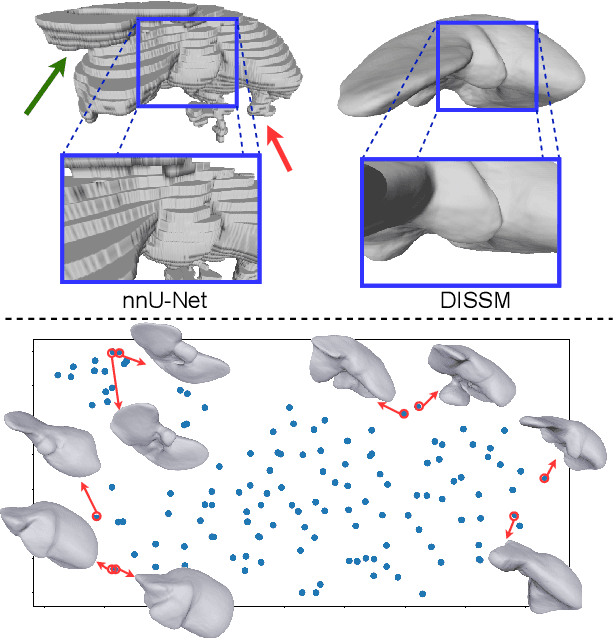
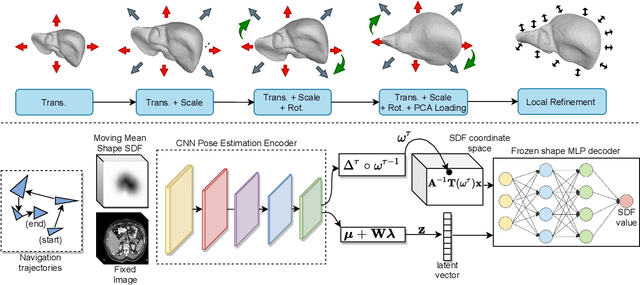
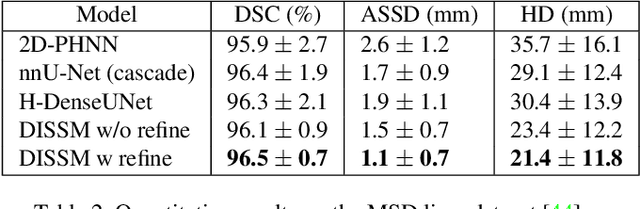
Abstract:3D delineation of anatomical structures is a cardinal goal in medical imaging analysis. Prior to deep learning, statistical shape models that imposed anatomical constraints and produced high quality surfaces were a core technology. Prior to deep learning, statistical shape models that imposed anatomical constraints and produced high quality surfaces were a core technology. Today fully-convolutional networks (FCNs), while dominant, do not offer these capabilities. We present deep implicit statistical shape models (DISSMs), a new approach to delineation that marries the representation power of convolutional neural networks (CNNs) with the robustness of SSMs. DISSMs use a deep implicit surface representation to produce a compact and descriptive shape latent space that permits statistical models of anatomical variance. To reliably fit anatomically plausible shapes to an image, we introduce a novel rigid and non-rigid pose estimation pipeline that is modelled as a Markov decision process(MDP). We outline a training regime that includes inverted episodic training and a deep realization of marginal space learning (MSL). Intra-dataset experiments on the task of pathological liver segmentation demonstrate that DISSMs can perform more robustly than three leading FCN models, including nnU-Net: reducing the mean Hausdorff distance (HD) by 7.7-14.3mm and improving the worst case Dice-Sorensen coefficient (DSC) by 1.2-2.3%. More critically, cross-dataset experiments on a dataset directly reflecting clinical deployment scenarios demonstrate that DISSMs improve the mean DSC and HD by 3.5-5.9% and 12.3-24.5mm, respectively, and the worst-case DSC by 5.4-7.3%. These improvements are over and above any benefits from representing delineations with high-quality surface.
Hetero-Modal Learning and Expansive Consistency Constraints for Semi-Supervised Detection from Multi-Sequence Data
Mar 24, 2021



Abstract:Lesion detection serves a critical role in early diagnosis and has been well explored in recent years due to methodological advancesand increased data availability. However, the high costs of annotations hinder the collection of large and completely labeled datasets, motivating semi-supervised detection approaches. In this paper, we introduce mean teacher hetero-modal detection (MTHD), which addresses two important gaps in current semi-supervised detection. First, it is not obvious how to enforce unlabeled consistency constraints across the very different outputs of various detectors, which has resulted in various compromises being used in the state of the art. Using an anchor-free framework, MTHD formulates a mean teacher approach without such compromises, enforcing consistency on the soft-output of object centers and size. Second, multi-sequence data is often critical, e.g., for abdominal lesion detection, but unlabeled data is often missing sequences. To deal with this, MTHD incorporates hetero-modal learning in its framework. Unlike prior art, MTHD is able to incorporate an expansive set of consistency constraints that include geometric transforms and random sequence combinations. We train and evaluate MTHD on liver lesion detection using the largest MR lesion dataset to date (1099 patients with >5000 volumes). MTHD surpasses the best fully-supervised and semi-supervised competitors by 10.1% and 3.5%, respectively, in average sensitivity.
A New Window Loss Function for Bone Fracture Detection and Localization in X-ray Images with Point-based Annotation
Jan 04, 2021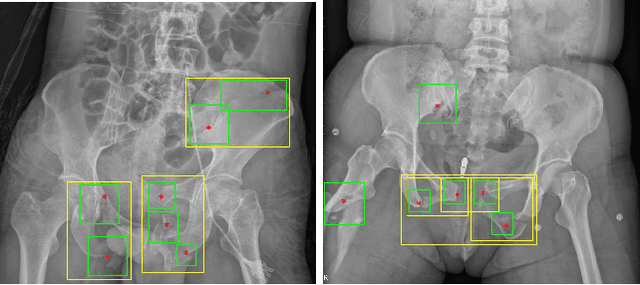
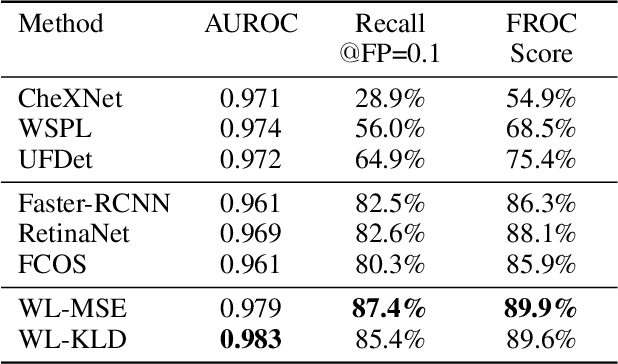
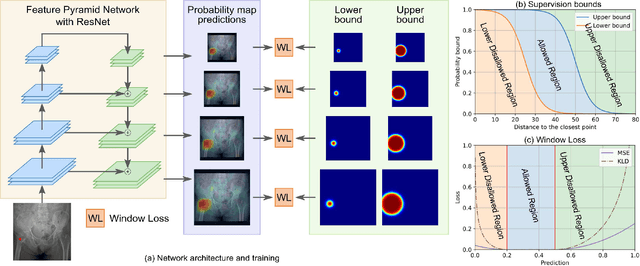
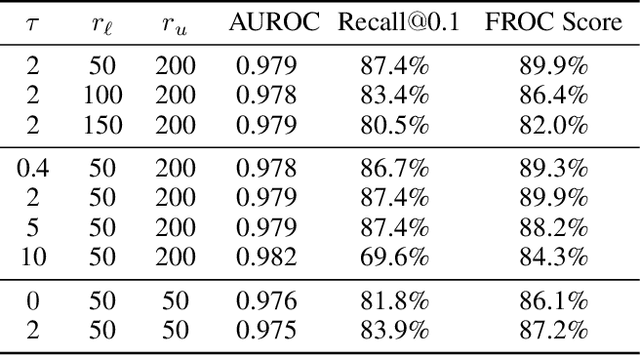
Abstract:Object detection methods are widely adopted for computer-aided diagnosis using medical images. Anomalous findings are usually treated as objects that are described by bounding boxes. Yet, many pathological findings, e.g., bone fractures, cannot be clearly defined by bounding boxes, owing to considerable instance, shape and boundary ambiguities. This makes bounding box annotations, and their associated losses, highly ill-suited. In this work, we propose a new bone fracture detection method for X-ray images, based on a labor effective and flexible annotation scheme suitable for abnormal findings with no clear object-level spatial extents or boundaries. Our method employs a simple, intuitive, and informative point-based annotation protocol to mark localized pathology information. To address the uncertainty in the fracture scales annotated via point(s), we convert the annotations into pixel-wise supervision that uses lower and upper bounds with positive, negative, and uncertain regions. A novel Window Loss is subsequently proposed to only penalize the predictions outside of the uncertain regions. Our method has been extensively evaluated on 4410 pelvic X-ray images of unique patients. Experiments demonstrate that our method outperforms previous state-of-the-art image classification and object detection baselines by healthy margins, with an AUROC of 0.983 and FROC score of 89.6%.
Deep Lesion Tracker: Monitoring Lesions in 4D Longitudinal Imaging Studies
Dec 09, 2020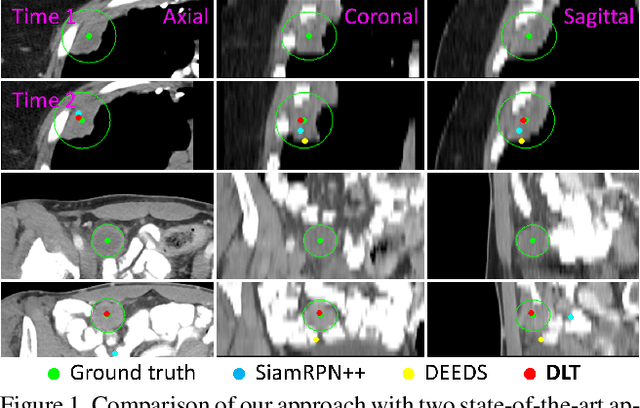
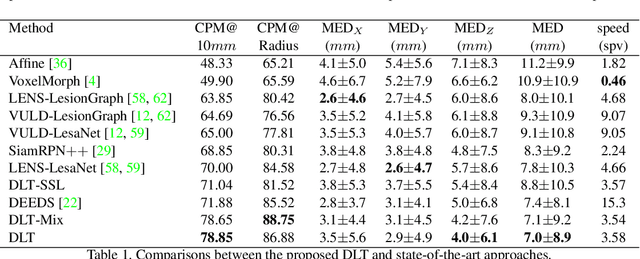
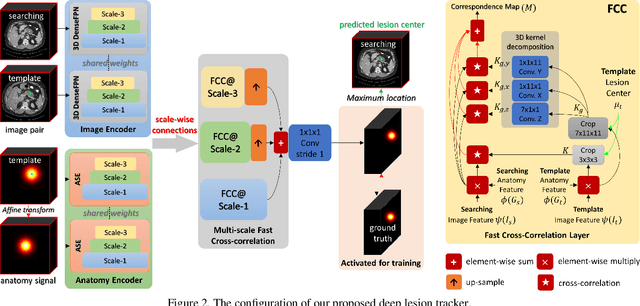
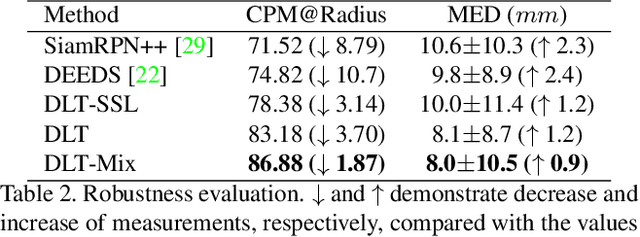
Abstract:Monitoring treatment response in longitudinal studies plays an important role in clinical practice. Accurately identifying lesions across serial imaging follow-up is the core to the monitoring procedure. Typically this incorporates both image and anatomical considerations. However, matching lesions manually is labor-intensive and time-consuming. In this work, we present deep lesion tracker (DLT), a deep learning approach that uses both appearance- and anatomical-based signals. To incorporate anatomical constraints, we propose an anatomical signal encoder, which prevents lesions being matched with visually similar but spurious regions. In addition, we present a new formulation for Siamese networks that avoids the heavy computational loads of 3D cross-correlation. To present our network with greater varieties of images, we also propose a self-supervised learning (SSL) strategy to train trackers with unpaired images, overcoming barriers to data collection. To train and evaluate our tracker, we introduce and release the first lesion tracking benchmark, consisting of 3891 lesion pairs from the public DeepLesion database. The proposed method, DLT, locates lesion centers with a mean error distance of 7 mm. This is 5% better than a leading registration algorithm while running 14 times faster on whole CT volumes. We demonstrate even greater improvements over detector or similarity-learning alternatives. DLT also generalizes well on an external clinical test set of 100 longitudinal studies, achieving 88% accuracy. Finally, we plug DLT into an automatic tumor monitoring workflow where it leads to an accuracy of 85% in assessing lesion treatment responses, which is only 0.46% lower than the accuracy of manual inputs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge