Chien-Hung Liao
A Cascaded Approach for ultraly High Performance Lesion Detection and False Positive Removal in Liver CT Scans
Jun 28, 2023Abstract:Liver cancer has high morbidity and mortality rates in the world. Multi-phase CT is a main medical imaging modality for detecting/identifying and diagnosing liver tumors. Automatically detecting and classifying liver lesions in CT images have the potential to improve the clinical workflow. This task remains challenging due to liver lesions' large variations in size, appearance, image contrast, and the complexities of tumor types or subtypes. In this work, we customize a multi-object labeling tool for multi-phase CT images, which is used to curate a large-scale dataset containing 1,631 patients with four-phase CT images, multi-organ masks, and multi-lesion (six major types of liver lesions confirmed by pathology) masks. We develop a two-stage liver lesion detection pipeline, where the high-sensitivity detecting algorithms in the first stage discover as many lesion proposals as possible, and the lesion-reclassification algorithms in the second stage remove as many false alarms as possible. The multi-sensitivity lesion detection algorithm maximizes the information utilization of the individual probability maps of segmentation, and the lesion-shuffle augmentation effectively explores the texture contrast between lesions and the liver. Independently tested on 331 patient cases, the proposed model achieves high sensitivity and specificity for malignancy classification in the multi-phase contrast-enhanced CT (99.2%, 97.1%, diagnosis setting) and in the noncontrast CT (97.3%, 95.7%, screening setting).
A Flexible Three-Dimensional Hetero-phase Computed Tomography Hepatocellular Carcinoma (HCC) Detection Algorithm for Generalizable and Practical HCC Screening
Aug 17, 2021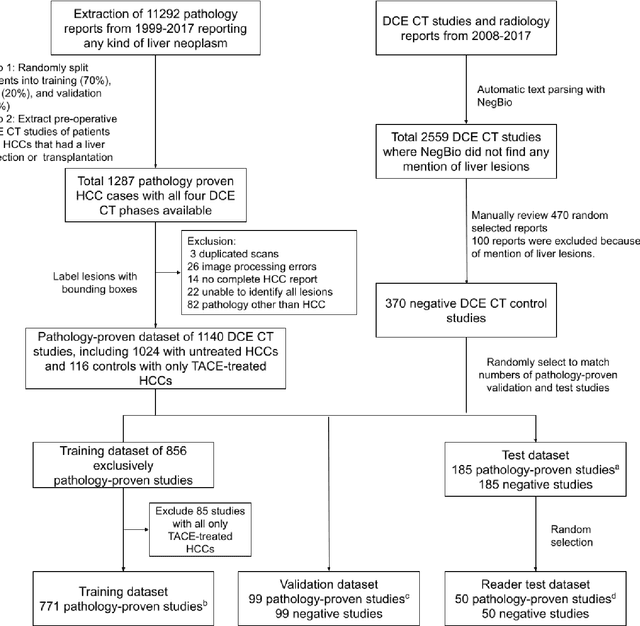
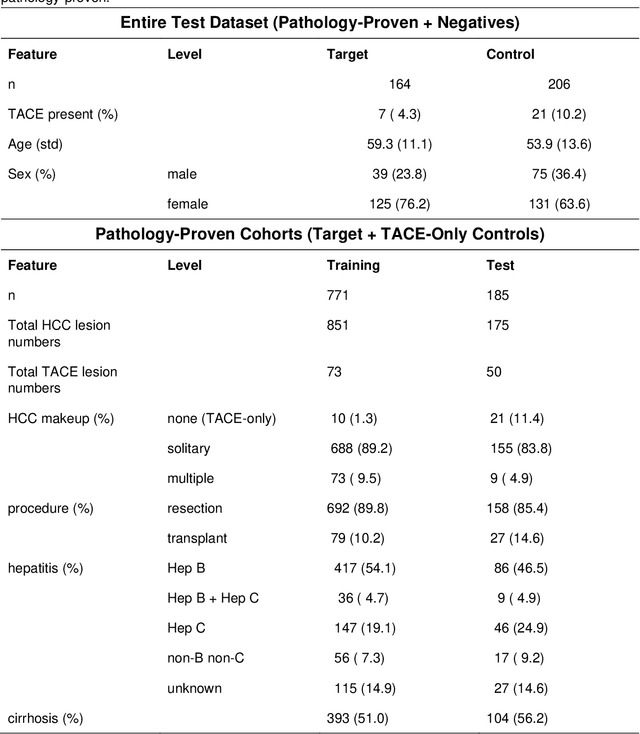
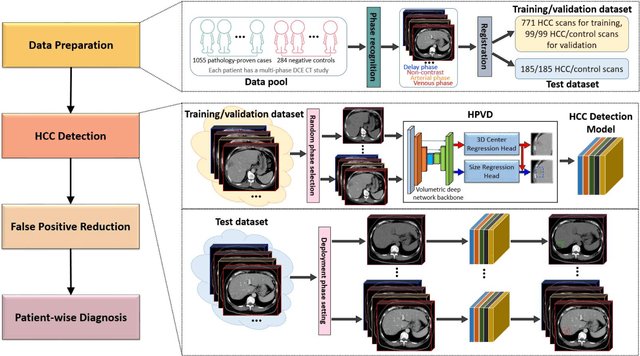
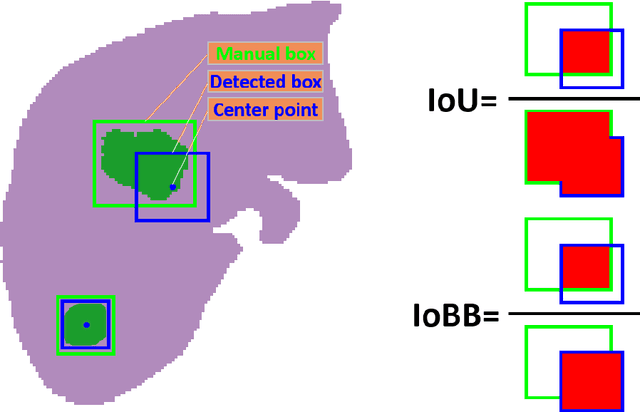
Abstract:Hepatocellular carcinoma (HCC) can be potentially discovered from abdominal computed tomography (CT) studies under varied clinical scenarios, e.g., fully dynamic contrast enhanced (DCE) studies, non-contrast (NC) plus venous phase (VP) abdominal studies, or NC-only studies. We develop a flexible three-dimensional deep algorithm, called hetero-phase volumetric detection (HPVD), that can accept any combination of contrast-phase inputs and with adjustable sensitivity depending on the clinical purpose. We trained HPVD on 771 DCE CT scans to detect HCCs and tested on external 164 positives and 206 controls, respectively. We compare performance against six clinical readers, including two radiologists, two hepato-pancreatico-biliary (HPB) surgeons, and two hepatologists. The area under curve (AUC) of the localization receiver operating characteristic (LROC) for NC-only, NC plus VP, and full DCE CT yielded 0.71, 0.81, 0.89 respectively. At a high sensitivity operating point of 80% on DCE CT, HPVD achieved 97% specificity, which is comparable to measured physician performance. We also demonstrate performance improvements over more typical and less flexible non hetero-phase detectors. Thus, we demonstrate that a single deep learning algorithm can be effectively applied to diverse HCC detection clinical scenarios.
Deep Implicit Statistical Shape Models for 3D Medical Image Delineation
Apr 07, 2021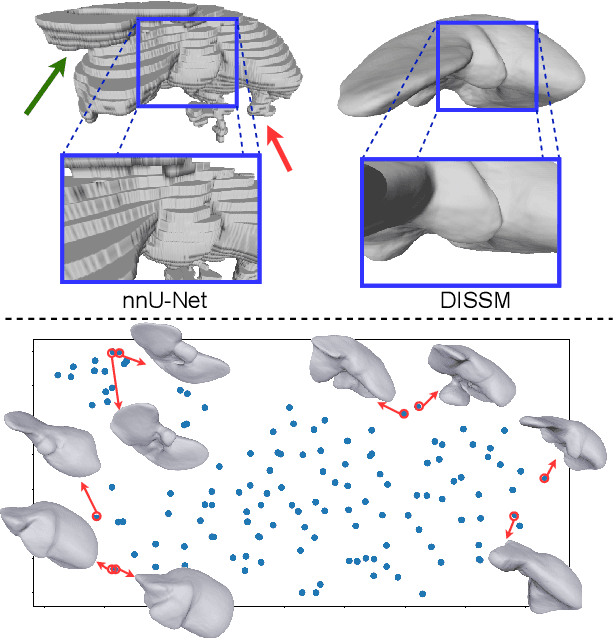
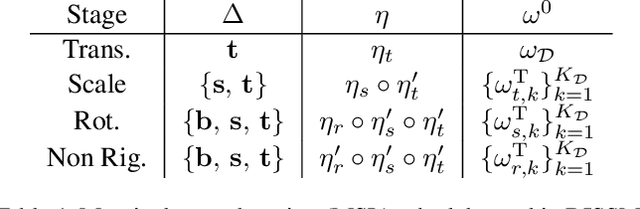
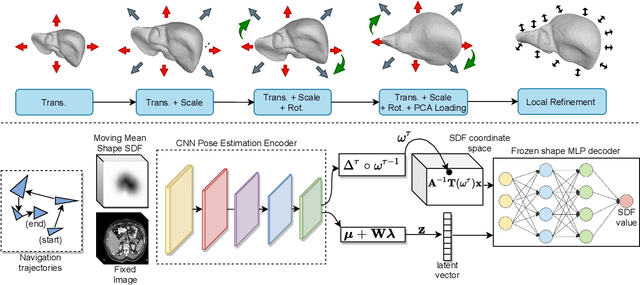
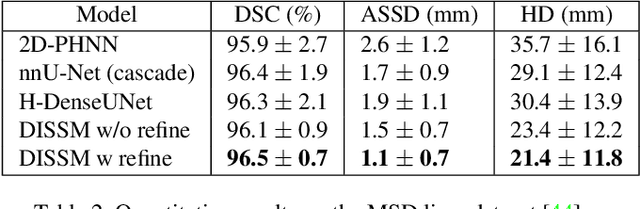
Abstract:3D delineation of anatomical structures is a cardinal goal in medical imaging analysis. Prior to deep learning, statistical shape models that imposed anatomical constraints and produced high quality surfaces were a core technology. Prior to deep learning, statistical shape models that imposed anatomical constraints and produced high quality surfaces were a core technology. Today fully-convolutional networks (FCNs), while dominant, do not offer these capabilities. We present deep implicit statistical shape models (DISSMs), a new approach to delineation that marries the representation power of convolutional neural networks (CNNs) with the robustness of SSMs. DISSMs use a deep implicit surface representation to produce a compact and descriptive shape latent space that permits statistical models of anatomical variance. To reliably fit anatomically plausible shapes to an image, we introduce a novel rigid and non-rigid pose estimation pipeline that is modelled as a Markov decision process(MDP). We outline a training regime that includes inverted episodic training and a deep realization of marginal space learning (MSL). Intra-dataset experiments on the task of pathological liver segmentation demonstrate that DISSMs can perform more robustly than three leading FCN models, including nnU-Net: reducing the mean Hausdorff distance (HD) by 7.7-14.3mm and improving the worst case Dice-Sorensen coefficient (DSC) by 1.2-2.3%. More critically, cross-dataset experiments on a dataset directly reflecting clinical deployment scenarios demonstrate that DISSMs improve the mean DSC and HD by 3.5-5.9% and 12.3-24.5mm, respectively, and the worst-case DSC by 5.4-7.3%. These improvements are over and above any benefits from representing delineations with high-quality surface.
A Practical Framework for ROI Detection in Medical Images -- a case study for hip detection in anteroposterior pelvic radiographs
Mar 02, 2021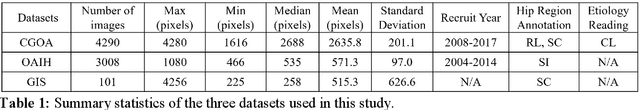
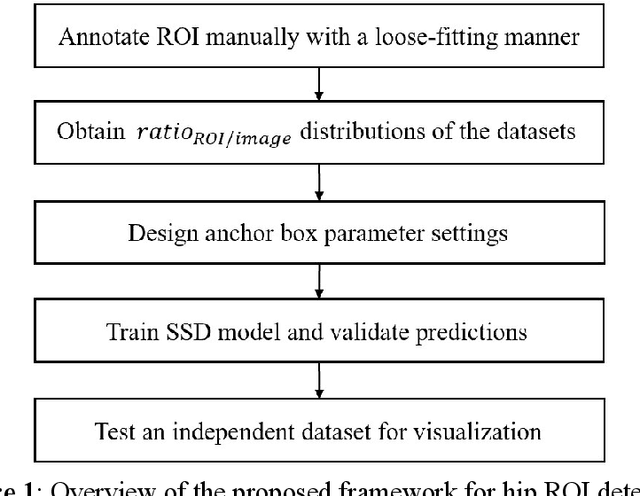
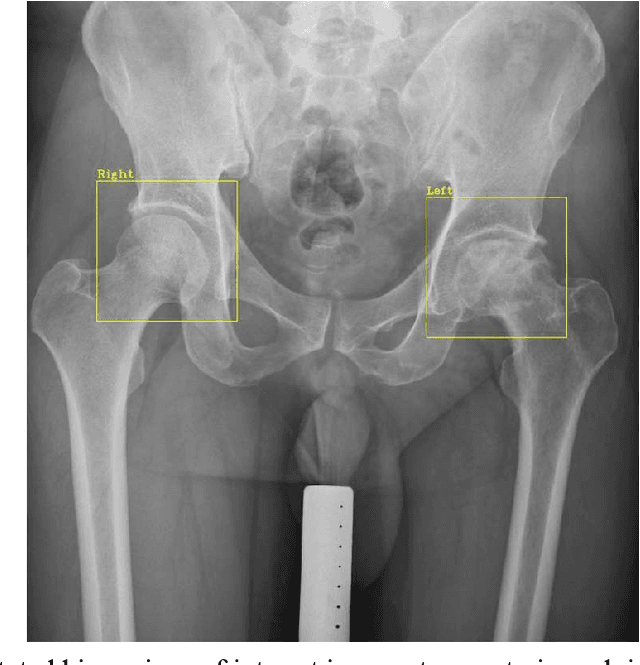
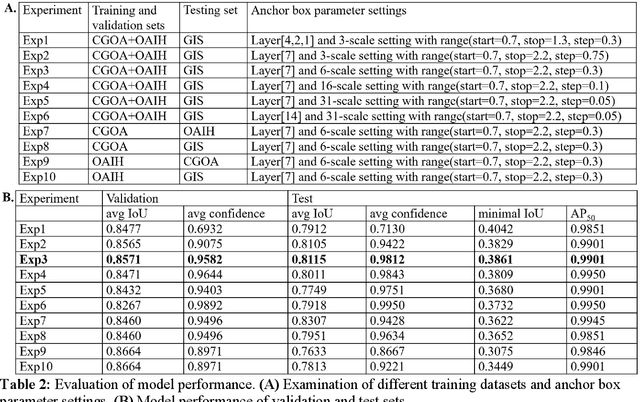
Abstract:Purpose Automated detection of region of interest (ROI) is a critical step for many medical image applications such as heart ROIs detection in perfusion MRI images, lung boundary detection in chest X-rays, and femoral head detection in pelvic radiographs. Thus, we proposed a practical framework of ROIs detection in medical images, with a case study for hip detection in anteroposterior (AP) pelvic radiographs. Materials and Methods: We conducted a retrospective study which analyzed hip joints seen on 7,399 AP pelvic radiographs from three diverse sources, including 4,290 high resolution radiographs from Chang Gung Memorial Hospital Osteoarthritis, 3,008 low to medium resolution radiographs from Osteoarthritis Initiative, and 101 heterogeneous radiographs from Google image search engine. We presented a deep learning-based ROI detection framework utilizing single-shot multi-box detector (SSD) with ResNet-101 backbone and customized head structure based on the characteristics of the obtained datasets, whose ground truths were labeled by non-medical annotators in a simple graphical interface. Results: Our method achieved average intersection over union (IoU)=0.8115, average confidence=0.9812, and average precision with threshold IoU=0.5 (AP50)=0.9901 in the independent test set, suggesting that the detected hip regions have appropriately covered main features of the hip joints. Conclusion: The proposed approach featured on low-cost labeling, data-driven model design, and heterogeneous data testing. We have demonstrated the feasibility of training a robust hip region detector for AP pelvic radiographs. This practical framework has a promising potential for a wide range of medical image applications.
A New Window Loss Function for Bone Fracture Detection and Localization in X-ray Images with Point-based Annotation
Jan 04, 2021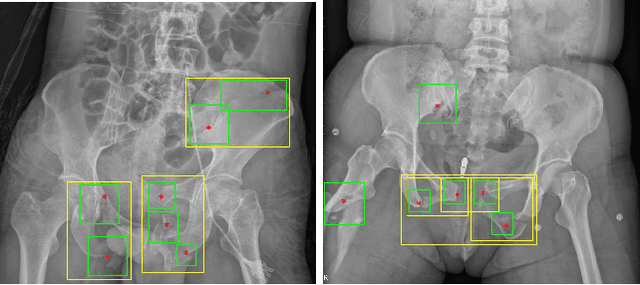
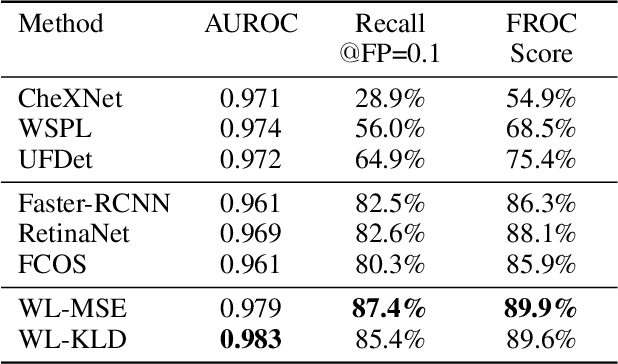
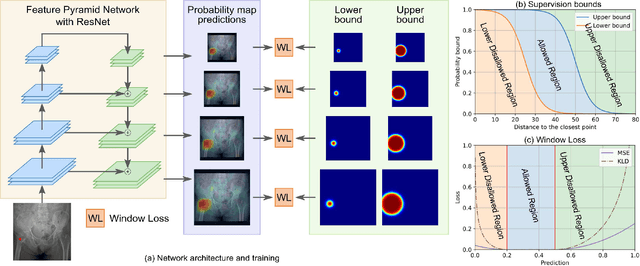
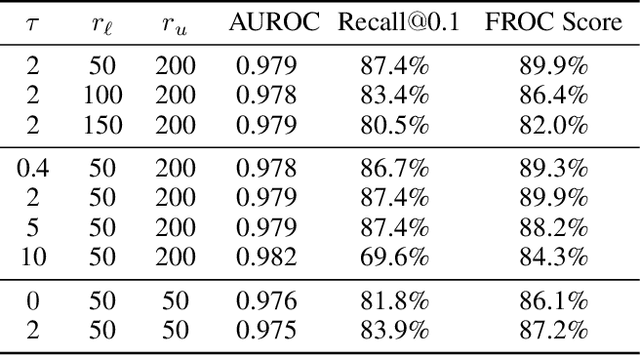
Abstract:Object detection methods are widely adopted for computer-aided diagnosis using medical images. Anomalous findings are usually treated as objects that are described by bounding boxes. Yet, many pathological findings, e.g., bone fractures, cannot be clearly defined by bounding boxes, owing to considerable instance, shape and boundary ambiguities. This makes bounding box annotations, and their associated losses, highly ill-suited. In this work, we propose a new bone fracture detection method for X-ray images, based on a labor effective and flexible annotation scheme suitable for abnormal findings with no clear object-level spatial extents or boundaries. Our method employs a simple, intuitive, and informative point-based annotation protocol to mark localized pathology information. To address the uncertainty in the fracture scales annotated via point(s), we convert the annotations into pixel-wise supervision that uses lower and upper bounds with positive, negative, and uncertain regions. A novel Window Loss is subsequently proposed to only penalize the predictions outside of the uncertain regions. Our method has been extensively evaluated on 4410 pelvic X-ray images of unique patients. Experiments demonstrate that our method outperforms previous state-of-the-art image classification and object detection baselines by healthy margins, with an AUROC of 0.983 and FROC score of 89.6%.
Knowledge Distillation with Adaptive Asymmetric Label Sharpening for Semi-supervised Fracture Detection in Chest X-rays
Dec 30, 2020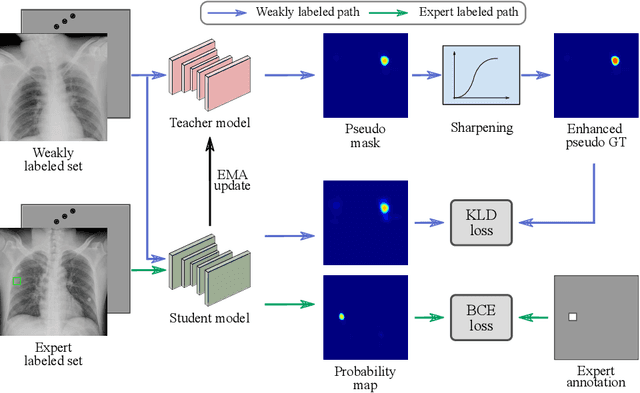
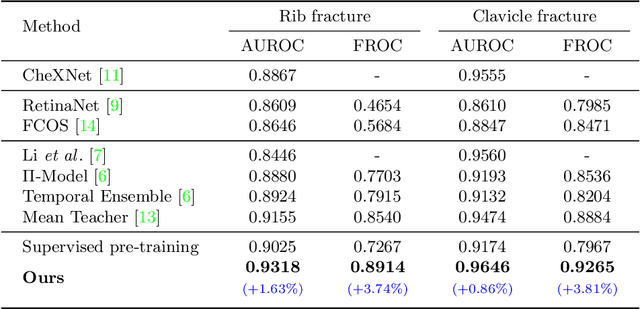
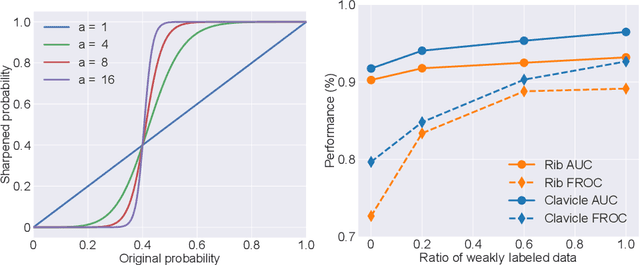
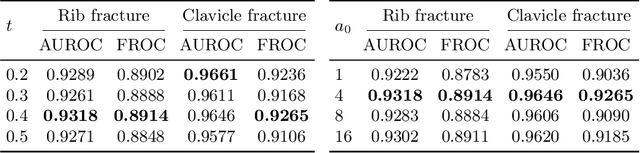
Abstract:Exploiting available medical records to train high performance computer-aided diagnosis (CAD) models via the semi-supervised learning (SSL) setting is emerging to tackle the prohibitively high labor costs involved in large-scale medical image annotations. Despite the extensive attentions received on SSL, previous methods failed to 1) account for the low disease prevalence in medical records and 2) utilize the image-level diagnosis indicated from the medical records. Both issues are unique to SSL for CAD models. In this work, we propose a new knowledge distillation method that effectively exploits large-scale image-level labels extracted from the medical records, augmented with limited expert annotated region-level labels, to train a rib and clavicle fracture CAD model for chest X-ray (CXR). Our method leverages the teacher-student model paradigm and features a novel adaptive asymmetric label sharpening (AALS) algorithm to address the label imbalance problem that specially exists in medical domain. Our approach is extensively evaluated on all CXR (N = 65,845) from the trauma registry of anonymous hospital over a period of 9 years (2008-2016), on the most common rib and clavicle fractures. The experiment results demonstrate that our method achieves the state-of-the-art fracture detection performance, i.e., an area under receiver operating characteristic curve (AUROC) of 0.9318 and a free-response receiver operating characteristic (FROC) score of 0.8914 on the rib fractures, significantly outperforming previous approaches by an AUROC gap of 1.63% and an FROC improvement by 3.74%. Consistent performance gains are also observed for clavicle fracture detection.
Deep Volumetric Universal Lesion Detection using Light-Weight Pseudo 3D Convolution and Surface Point Regression
Aug 30, 2020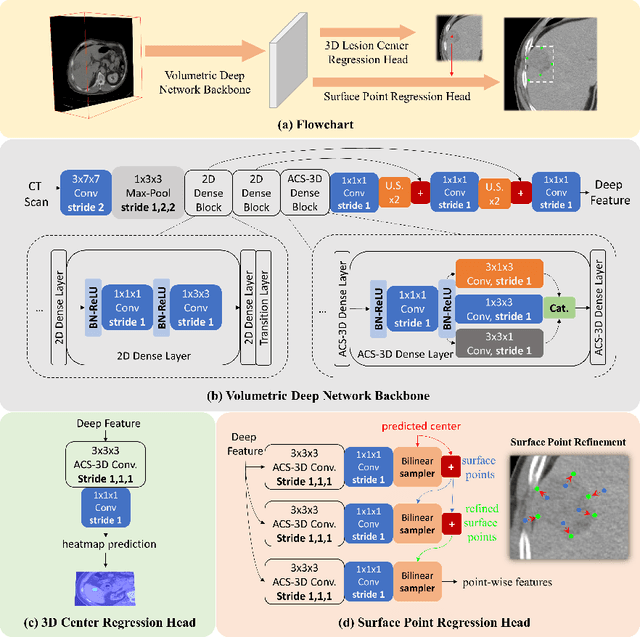
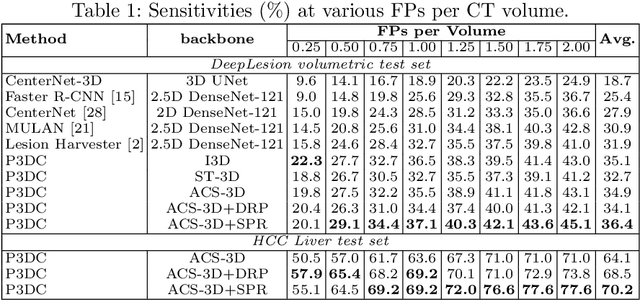
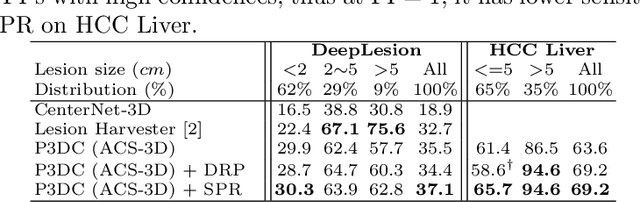
Abstract:Identifying, measuring and reporting lesions accurately and comprehensively from patient CT scans are important yet time-consuming procedures for physicians. Computer-aided lesion/significant-findings detection techniques are at the core of medical imaging, which remain very challenging due to the tremendously large variability of lesion appearance, location and size distributions in 3D imaging. In this work, we propose a novel deep anchor-free one-stage VULD framework that incorporates (1) P3DC operators to recycle the architectural configurations and pre-trained weights from the off-the-shelf 2D networks, especially ones with large capacities to cope with data variance, and (2) a new SPR method to effectively regress the 3D lesion spatial extents by pinpointing their representative key points on lesion surfaces. Experimental validations are first conducted on the public large-scale NIH DeepLesion dataset where our proposed method delivers new state-of-the-art quantitative performance. We also test VULD on our in-house dataset for liver tumor detection. VULD generalizes well in both large-scale and small-sized tumor datasets in CT imaging.
Anatomy-Aware Siamese Network: Exploiting Semantic Asymmetry for Accurate Pelvic Fracture Detection in X-ray Images
Jul 12, 2020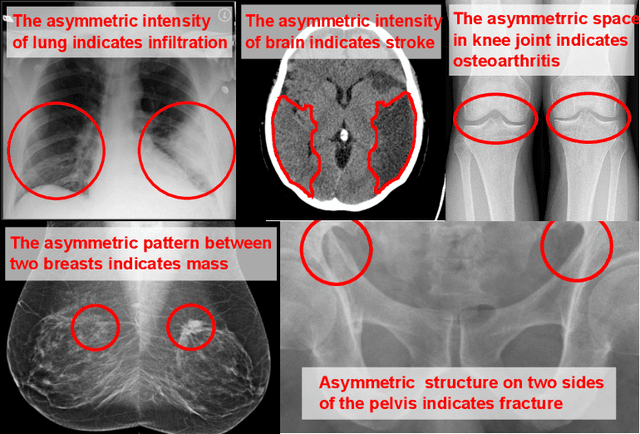
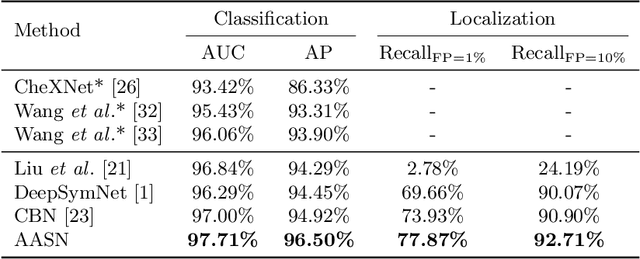
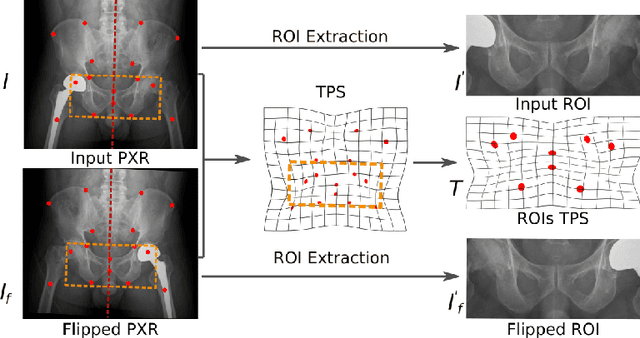
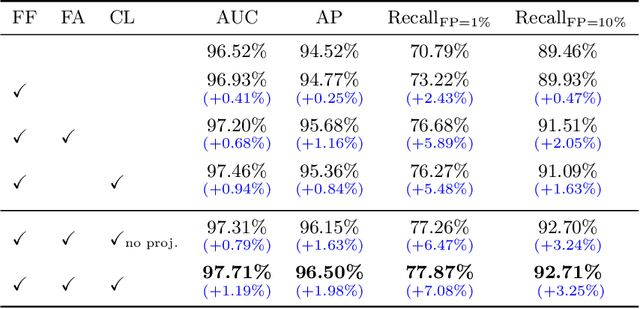
Abstract:Visual cues of enforcing bilaterally symmetric anatomies as normal findings are widely used in clinical practice to disambiguate subtle abnormalities from medical images. So far, inadequate research attention has been received on effectively emulating this practice in CAD methods. In this work, we exploit semantic anatomical symmetry or asymmetry analysis in a complex CAD scenario, i.e., anterior pelvic fracture detection in trauma PXRs, where semantically pathological (refer to as fracture) and non-pathological (e.g., pose) asymmetries both occur. Visually subtle yet pathologically critical fracture sites can be missed even by experienced clinicians, when limited diagnosis time is permitted in emergency care. We propose a novel fracture detection framework that builds upon a Siamese network enhanced with a spatial transformer layer to holistically analyze symmetric image features. Image features are spatially formatted to encode bilaterally symmetric anatomies. A new contrastive feature learning component in our Siamese network is designed to optimize the deep image features being more salient corresponding to the underlying semantic asymmetries (caused by pelvic fracture occurrences). Our proposed method have been extensively evaluated on 2,359 PXRs from unique patients (the largest study to-date), and report an area under ROC curve score of 0.9771. This is the highest among state-of-the-art fracture detection methods, with improved clinical indications.
Harvesting, Detecting, and Characterizing Liver Lesions from Large-scale Multi-phase CT Data via Deep Dynamic Texture Learning
Jun 28, 2020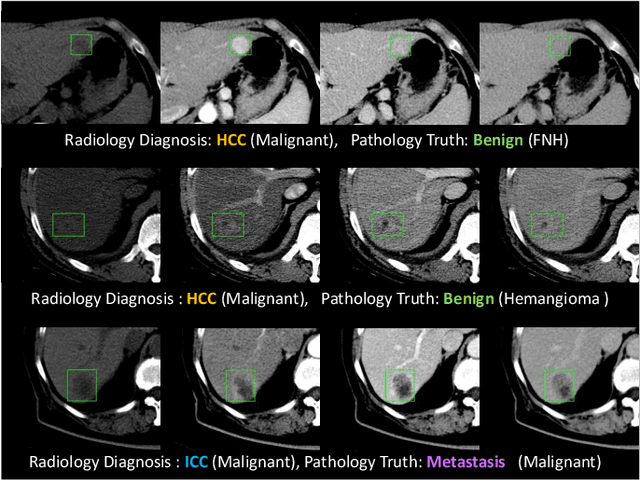
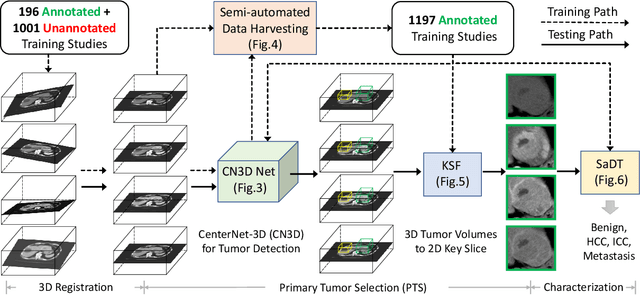
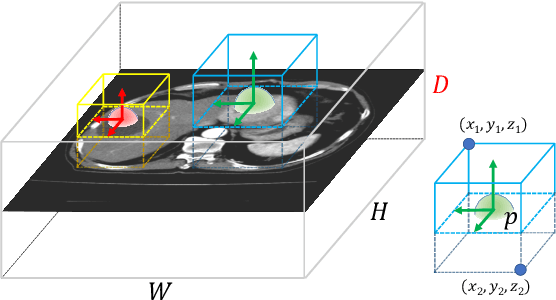
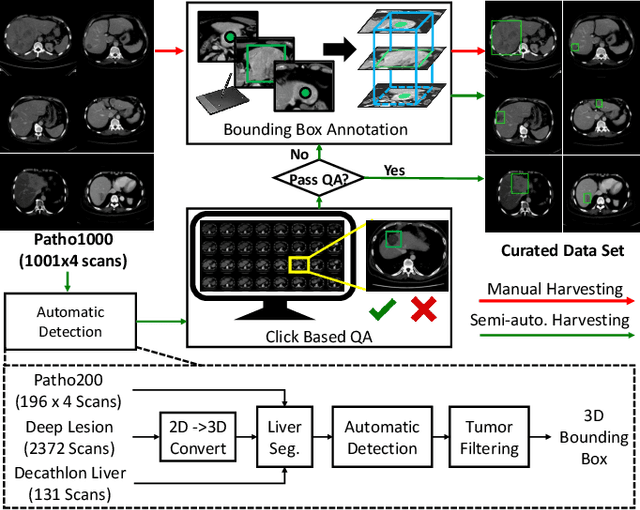
Abstract:Effective and non-invasive radiological imaging based tumor/lesion characterization (e.g., subtype classification) has long been a major aim in the oncology diagnosis and treatment procedures, with the hope of reducing needs for invasive surgical biopsies. Prior work are generally very restricted to a limited patient sample size, especially using patient studies with confirmed pathological reports as ground truth. In this work, we curate a patient cohort of 1305 dynamic contrast CT studies (i.e., 5220 multi-phase 3D volumes) with pathology confirmed ground truth. A novel fully-automated and multi-stage liver tumor characterization framework is proposed, comprising four steps of tumor proposal detection, tumor harvesting, primary tumor site selection, and deep texture-based characterization. More specifically, (1) we propose a 3D non-isotropic anchor-free lesion detection method; (2) we present and validate the use of multi-phase deep texture learning for precise liver lesion tissue characterization, named spatially adaptive deep texture (SaDT); (3) we leverage small-sized public datasets to semi-automatically curate our large-scale clinical dataset of 1305 patients where four main liver tumor subtypes of primary, secondary, metastasized and benign are presented. Extensive evaluations demonstrate that our new data curation strategy, combined with the SaDT deep dynamic texture analysis, can effectively improve the mean F1 scores by >8.6% compared with baselines, in differentiating four major liver lesion types. This is a significant step towards the clinical goal.
CT Data Curation for Liver Patients: Phase Recognition in Dynamic Contrast-Enhanced CT
Sep 27, 2019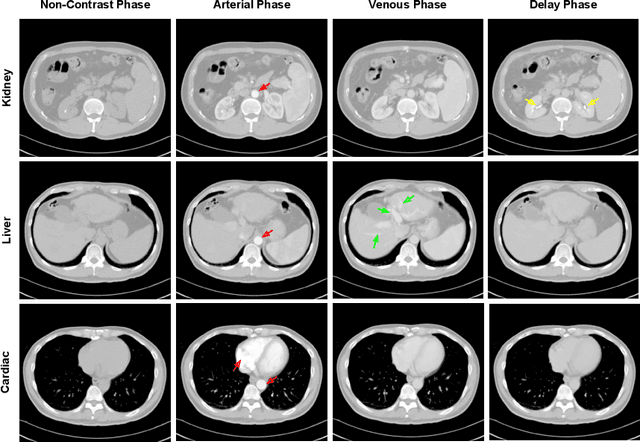
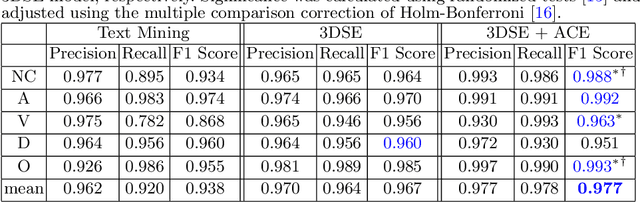
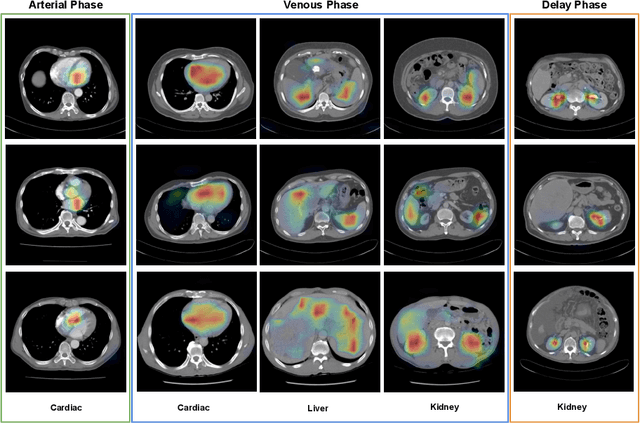
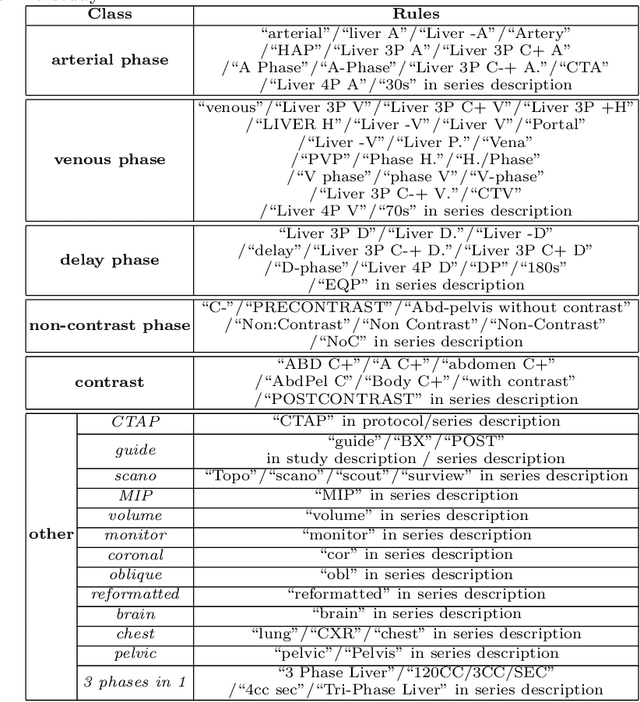
Abstract:As the demand for more descriptive machine learning models grows within medical imaging, bottlenecks due to data paucity will exacerbate. Thus, collecting enough large-scale data will require automated tools to harvest data/label pairs from messy and real-world datasets, such as hospital PACS. This is the focus of our work, where we present a principled data curation tool to extract multi-phase CT liver studies and identify each scan's phase from a real-world and heterogenous hospital PACS dataset. Emulating a typical deployment scenario, we first obtain a set of noisy labels from our institutional partners that are text mined using simple rules from DICOM tags. We train a deep learning system, using a customized and streamlined 3D SE architecture, to identify non-contrast, arterial, venous, and delay phase dynamic CT liver scans, filtering out anything else, including other types of liver contrast studies. To exploit as much training data as possible, we also introduce an aggregated cross entropy loss that can learn from scans only identified as "contrast". Extensive experiments on a dataset of 43K scans of 7680 patient imaging studies demonstrate that our 3DSE architecture, armed with our aggregated loss, can achieve a mean F1 of 0.977 and can correctly harvest up to 92.7% of studies, which significantly outperforms the text-mined and standard-loss approach, and also outperforms other, and more complex, model architectures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge