Youjing Zheng
A Flexible Three-Dimensional Hetero-phase Computed Tomography Hepatocellular Carcinoma (HCC) Detection Algorithm for Generalizable and Practical HCC Screening
Aug 17, 2021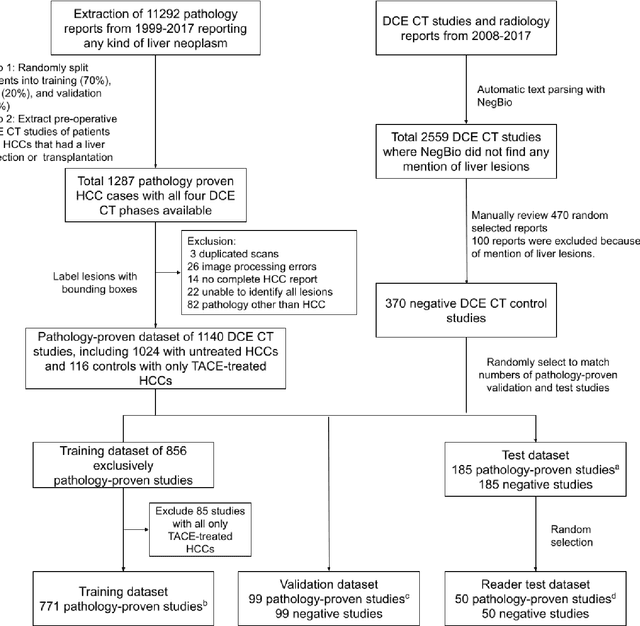
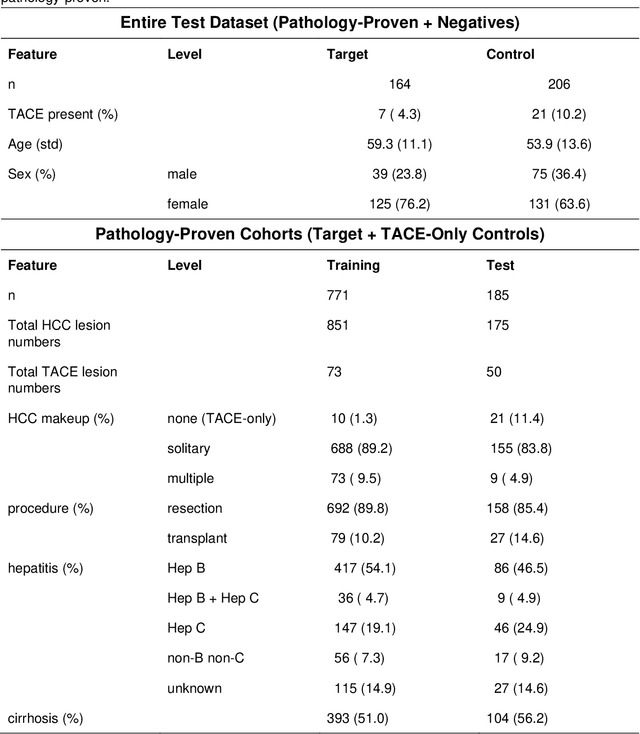
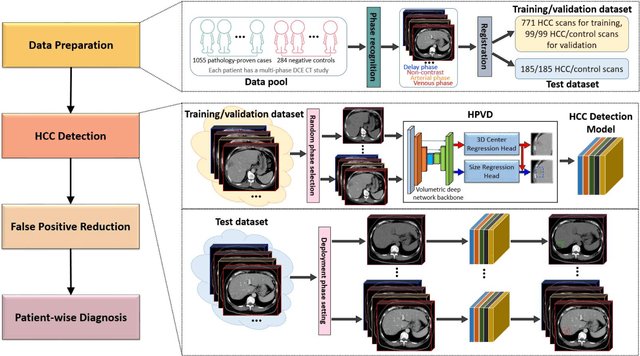
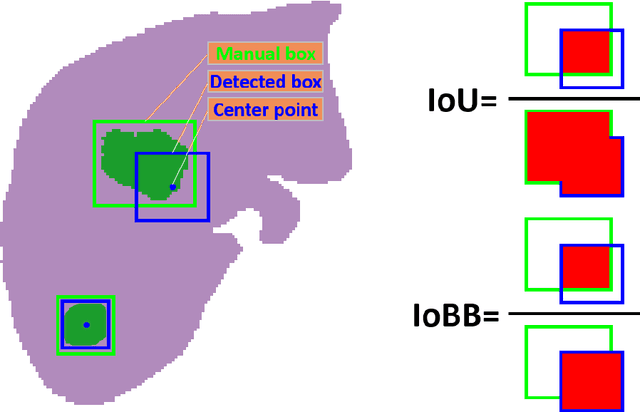
Abstract:Hepatocellular carcinoma (HCC) can be potentially discovered from abdominal computed tomography (CT) studies under varied clinical scenarios, e.g., fully dynamic contrast enhanced (DCE) studies, non-contrast (NC) plus venous phase (VP) abdominal studies, or NC-only studies. We develop a flexible three-dimensional deep algorithm, called hetero-phase volumetric detection (HPVD), that can accept any combination of contrast-phase inputs and with adjustable sensitivity depending on the clinical purpose. We trained HPVD on 771 DCE CT scans to detect HCCs and tested on external 164 positives and 206 controls, respectively. We compare performance against six clinical readers, including two radiologists, two hepato-pancreatico-biliary (HPB) surgeons, and two hepatologists. The area under curve (AUC) of the localization receiver operating characteristic (LROC) for NC-only, NC plus VP, and full DCE CT yielded 0.71, 0.81, 0.89 respectively. At a high sensitivity operating point of 80% on DCE CT, HPVD achieved 97% specificity, which is comparable to measured physician performance. We also demonstrate performance improvements over more typical and less flexible non hetero-phase detectors. Thus, we demonstrate that a single deep learning algorithm can be effectively applied to diverse HCC detection clinical scenarios.
Learning from Multiple Datasets with Heterogeneous and Partial Labels for Universal Lesion Detection in CT
Sep 05, 2020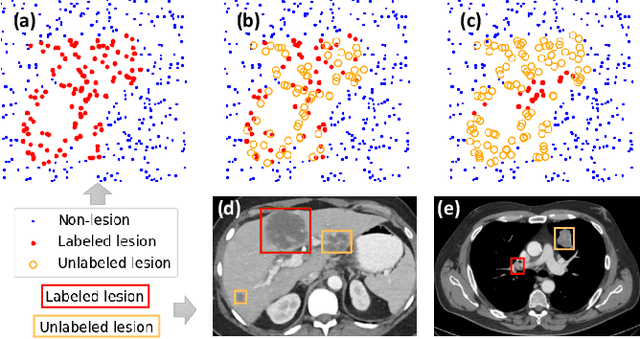
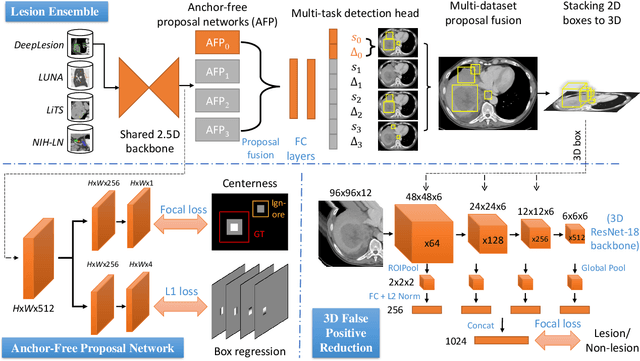
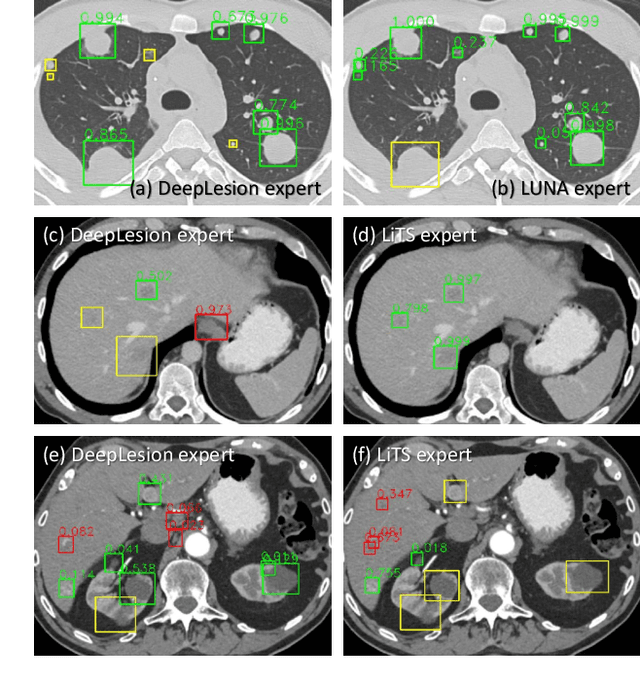

Abstract:Large-scale datasets with high-quality labels are desired for training accurate deep learning models. However, due to annotation costs, medical imaging datasets are often either partially-labeled or small. For example, DeepLesion is a large-scale CT image dataset with lesions of various types, but it also has many unlabeled lesions (missing annotations). When training a lesion detector on a partially-labeled dataset, the missing annotations will generate incorrect negative signals and degrade performance. Besides DeepLesion, there are several small single-type datasets, such as LUNA for lung nodules and LiTS for liver tumors. Such datasets have heterogeneous label scopes, i.e., different lesion types are labeled in different datasets with other types ignored. In this work, we aim to tackle the problem of heterogeneous and partial labels, and develop a universal lesion detection algorithm to detect a comprehensive variety of lesions. First, we build a simple yet effective lesion detection framework named Lesion ENSemble (LENS). LENS can efficiently learn from multiple heterogeneous lesion datasets in a multi-task fashion and leverage their synergy by feature sharing and proposal fusion. Next, we propose strategies to mine missing annotations from partially-labeled datasets by exploiting clinical prior knowledge and cross-dataset knowledge transfer. Finally, we train our framework on four public lesion datasets and evaluate it on 800 manually-labeled sub-volumes in DeepLesion. On this challenging task, our method brings a relative improvement of 49% compared to the current state-of-the-art approach.
Lesion Harvester: Iteratively Mining Unlabeled Lesions and Hard-Negative Examples at Scale
Jan 28, 2020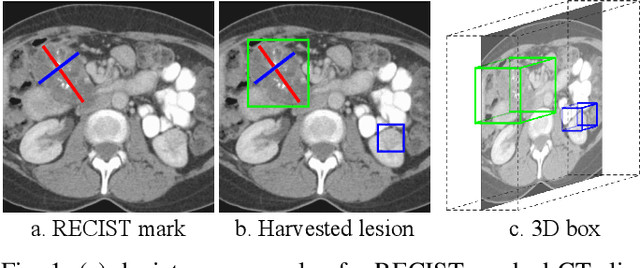
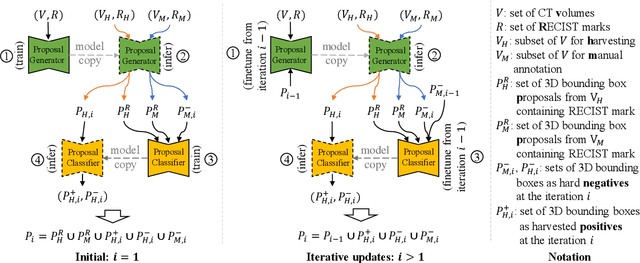
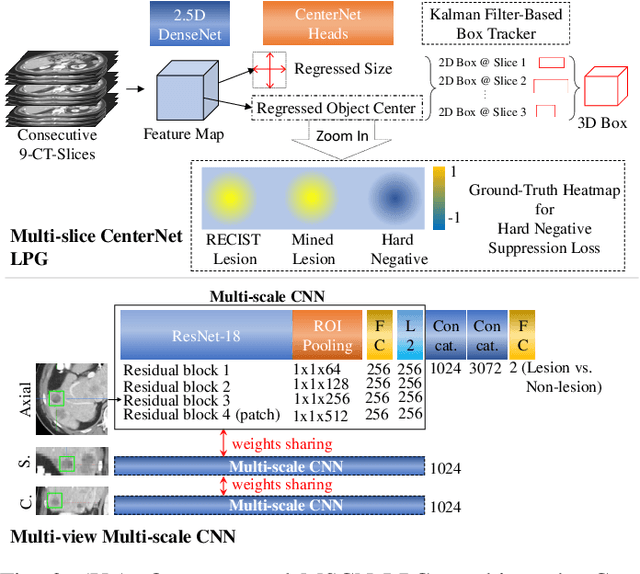
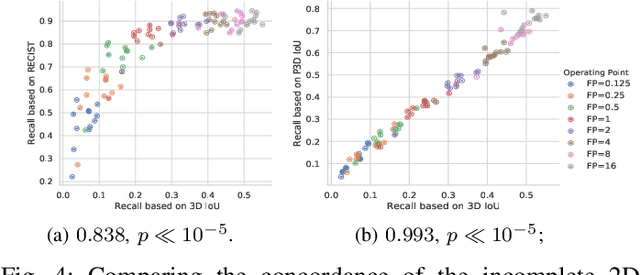
Abstract:Acquiring large-scale medical image data, necessary for training machine learning algorithms, is frequently intractable, due to prohibitive expert-driven annotation costs. Recent datasets extracted from hospital archives, e.g., DeepLesion, have begun to address this problem. However, these are often incompletely or noisily labeled, e.g., DeepLesion leaves over 50% of its lesions unlabeled. Thus, effective methods to harvest missing annotations are critical for continued progress in medical image analysis. This is the goal of our work, where we develop a powerful system to harvest missing lesions from the DeepLesion dataset at high precision. Accepting the need for some degree of expert labor to achieve high fidelity, we exploit a small fully-labeled subset of medical image volumes and use it to intelligently mine annotations from the remainder. To do this, we chain together a highly sensitive lesion proposal generator and a very selective lesion proposal classifier. While our framework is generic, we optimize our performance by proposing a 3D contextual lesion proposal generator and by using a multi-view multi-scale lesion proposal classifier. These produce harvested and hard-negative proposals, which we then re-use to finetune our proposal generator by using a novel hard negative suppression loss, continuing this process until no extra lesions are found. Extensive experimental analysis demonstrates that our method can harvest an additional 9,805 lesions while keeping precision above 90%. To demonstrate the benefits of our approach, we show that lesion detectors trained on our harvested lesions can significantly outperform the same variants only trained on the original annotations, with boost of average precision of 7% to 10%. We open source our annotations at https://github.com/JimmyCai91/DeepLesionAnnotation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge