Ashwin Raju
Inpainting Pathology in Lumbar Spine MRI with Latent Diffusion
Jun 04, 2024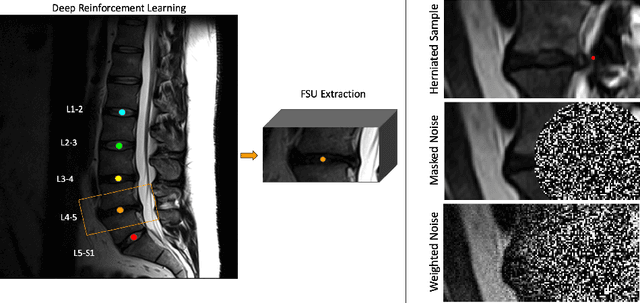
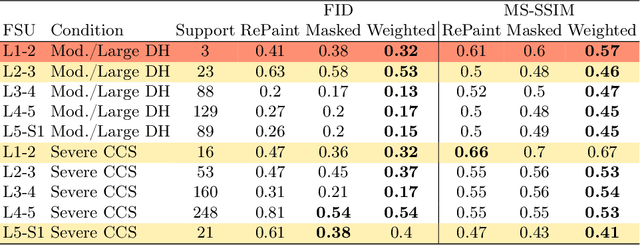
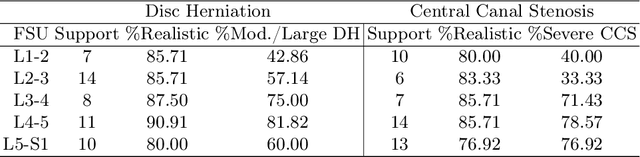
Abstract:Data driven models for automated diagnosis in radiology suffer from insufficient and imbalanced datasets due to low representation of pathology in a population and the cost of expert annotations. Datasets can be bolstered through data augmentation. However, even when utilizing a full suite of transformations during model training, typical data augmentations do not address variations in human anatomy. An alternative direction is to synthesize data using generative models, which can potentially craft datasets with specific attributes. While this holds promise, commonly used generative models such as Generative Adversarial Networks may inadvertently produce anatomically inaccurate features. On the other hand, diffusion models, which offer greater stability, tend to memorize training data, raising concerns about privacy and generative diversity. Alternatively, inpainting has the potential to augment data through directly inserting pathology in medical images. However, this approach introduces a new challenge: accurately merging the generated pathological features with the surrounding anatomical context. While inpainting is a well established method for addressing simple lesions, its application to pathologies that involve complex structural changes remains relatively unexplored. We propose an efficient method for inpainting pathological features onto healthy anatomy in MRI through voxelwise noise scheduling in a latent diffusion model. We evaluate the method's ability to insert disc herniation and central canal stenosis in lumbar spine sagittal T2 MRI, and it achieves superior Frechet Inception Distance compared to state-of-the-art methods.
Deep Implicit Statistical Shape Models for 3D Medical Image Delineation
Apr 07, 2021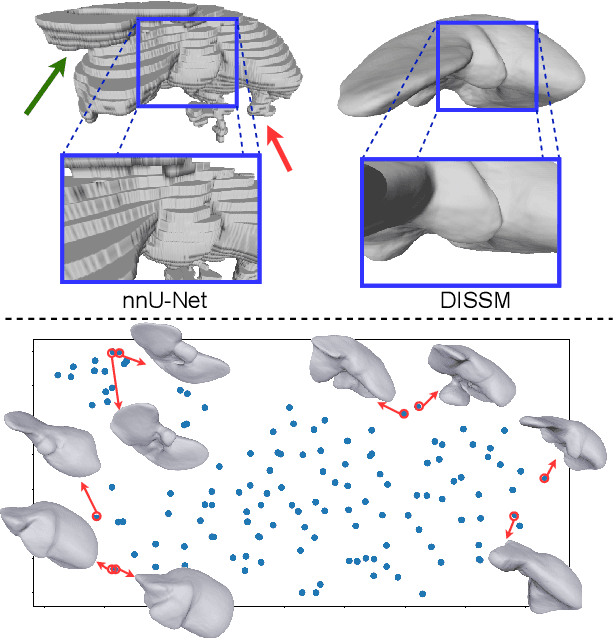
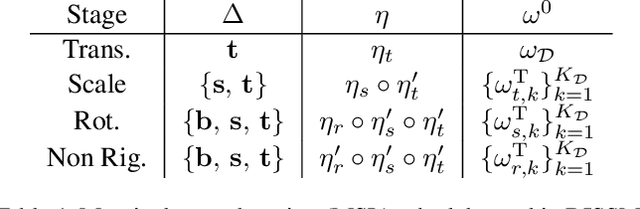
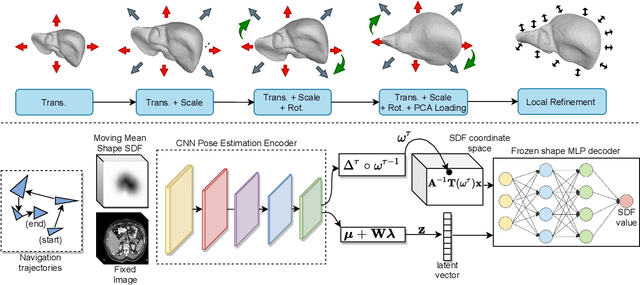
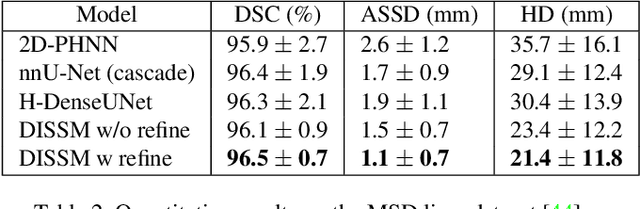
Abstract:3D delineation of anatomical structures is a cardinal goal in medical imaging analysis. Prior to deep learning, statistical shape models that imposed anatomical constraints and produced high quality surfaces were a core technology. Prior to deep learning, statistical shape models that imposed anatomical constraints and produced high quality surfaces were a core technology. Today fully-convolutional networks (FCNs), while dominant, do not offer these capabilities. We present deep implicit statistical shape models (DISSMs), a new approach to delineation that marries the representation power of convolutional neural networks (CNNs) with the robustness of SSMs. DISSMs use a deep implicit surface representation to produce a compact and descriptive shape latent space that permits statistical models of anatomical variance. To reliably fit anatomically plausible shapes to an image, we introduce a novel rigid and non-rigid pose estimation pipeline that is modelled as a Markov decision process(MDP). We outline a training regime that includes inverted episodic training and a deep realization of marginal space learning (MSL). Intra-dataset experiments on the task of pathological liver segmentation demonstrate that DISSMs can perform more robustly than three leading FCN models, including nnU-Net: reducing the mean Hausdorff distance (HD) by 7.7-14.3mm and improving the worst case Dice-Sorensen coefficient (DSC) by 1.2-2.3%. More critically, cross-dataset experiments on a dataset directly reflecting clinical deployment scenarios demonstrate that DISSMs improve the mean DSC and HD by 3.5-5.9% and 12.3-24.5mm, respectively, and the worst-case DSC by 5.4-7.3%. These improvements are over and above any benefits from representing delineations with high-quality surface.
User-Guided Domain Adaptation for Rapid Annotation from User Interactions: A Study on Pathological Liver Segmentation
Sep 05, 2020



Abstract:Mask-based annotation of medical images, especially for 3D data, is a bottleneck in developing reliable machine learning models. Using minimal-labor user interactions (UIs) to guide the annotation is promising, but challenges remain on best harmonizing the mask prediction with the UIs. To address this, we propose the user-guided domain adaptation (UGDA) framework, which uses prediction-based adversarial domain adaptation (PADA) to model the combined distribution of UIs and mask predictions. The UIs are then used as anchors to guide and align the mask prediction. Importantly, UGDA can both learn from unlabelled data and also model the high-level semantic meaning behind different UIs. We test UGDA on annotating pathological livers using a clinically comprehensive dataset of 927 patient studies. Using only extreme-point UIs, we achieve a mean (worst-case) performance of 96.1%(94.9%), compared to 93.0% (87.0%) for deep extreme points (DEXTR). Furthermore, we also show UGDA can retain this state-of-the-art performance even when only seeing a fraction of available UIs, demonstrating an ability for robust and reliable UI-guided segmentation with extremely minimal labor demands.
Harvesting, Detecting, and Characterizing Liver Lesions from Large-scale Multi-phase CT Data via Deep Dynamic Texture Learning
Jun 28, 2020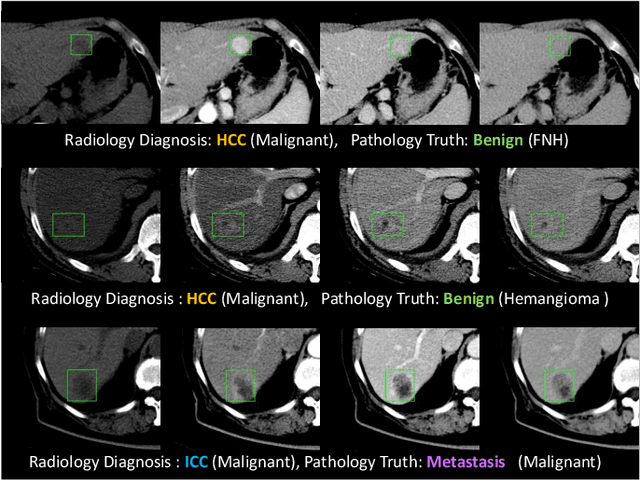
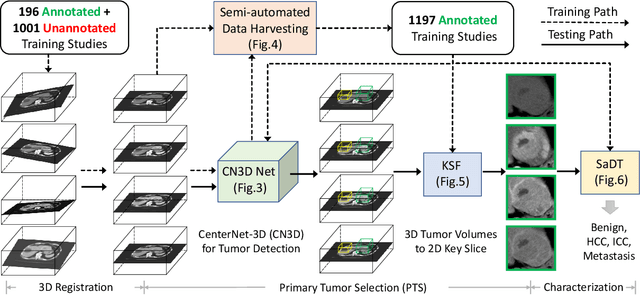
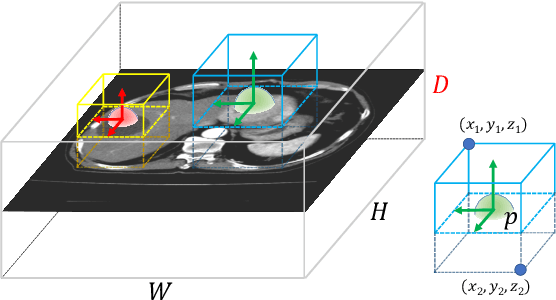
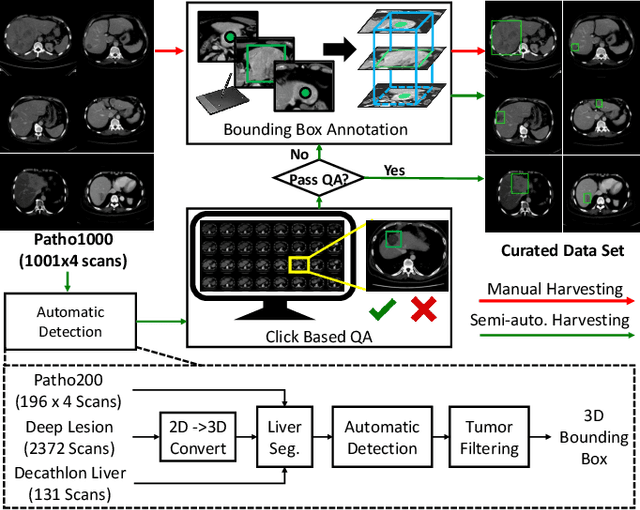
Abstract:Effective and non-invasive radiological imaging based tumor/lesion characterization (e.g., subtype classification) has long been a major aim in the oncology diagnosis and treatment procedures, with the hope of reducing needs for invasive surgical biopsies. Prior work are generally very restricted to a limited patient sample size, especially using patient studies with confirmed pathological reports as ground truth. In this work, we curate a patient cohort of 1305 dynamic contrast CT studies (i.e., 5220 multi-phase 3D volumes) with pathology confirmed ground truth. A novel fully-automated and multi-stage liver tumor characterization framework is proposed, comprising four steps of tumor proposal detection, tumor harvesting, primary tumor site selection, and deep texture-based characterization. More specifically, (1) we propose a 3D non-isotropic anchor-free lesion detection method; (2) we present and validate the use of multi-phase deep texture learning for precise liver lesion tissue characterization, named spatially adaptive deep texture (SaDT); (3) we leverage small-sized public datasets to semi-automatically curate our large-scale clinical dataset of 1305 patients where four main liver tumor subtypes of primary, secondary, metastasized and benign are presented. Extensive evaluations demonstrate that our new data curation strategy, combined with the SaDT deep dynamic texture analysis, can effectively improve the mean F1 scores by >8.6% compared with baselines, in differentiating four major liver lesion types. This is a significant step towards the clinical goal.
Co-Heterogeneous and Adaptive Segmentation from Multi-Source and Multi-Phase CT Imaging Data: A Study on Pathological Liver and Lesion Segmentation
Jun 10, 2020


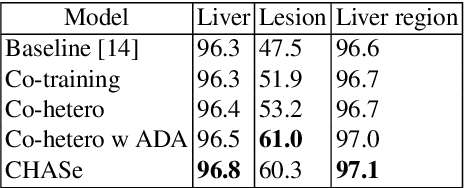
Abstract:In medical imaging, organ/pathology segmentation models trained on current publicly available and fully-annotated datasets usually do not well-represent the heterogeneous modalities, phases, pathologies, and clinical scenarios encountered in real environments. On the other hand, there are tremendous amounts of unlabelled patient imaging scans stored by many modern clinical centers. In this work, we present a novel segmentation strategy, co-heterogenous and adaptive segmentation (CHASe), which only requires a small labeled cohort of single phase imaging data to adapt to any unlabeled cohort of heterogenous multi-phase data with possibly new clinical scenarios and pathologies. To do this, we propose a versatile framework that fuses appearance based semi-supervision, mask based adversarial domain adaptation, and pseudo-labeling. We also introduce co-heterogeneous training, which is a novel integration of co-training and hetero modality learning. We have evaluated CHASe using a clinically comprehensive and challenging dataset of multi-phase computed tomography (CT) imaging studies (1147 patients and 4577 3D volumes). Compared to previous state-of-the-art baselines, CHASe can further improve pathological liver mask Dice-Sorensen coefficients by ranges of $4.2\% \sim 9.4\%$, depending on the phase combinations: e.g., from $84.6\%$ to $94.0\%$ on non-contrast CTs.
JSSR: A Joint Synthesis, Segmentation, and Registration System for 3D Multi-Modal Image Alignment of Large-scale Pathological CT Scans
May 27, 2020



Abstract:Multi-modal image registration is a challenging problem yet important clinical task in many real applications and scenarios. For medical imaging based diagnosis, deformable registration among different image modalities is often required in order to provide complementary visual information, as the first step. During the registration, the semantic information is the key to match homologous points and pixels. Nevertheless, many conventional registration methods are incapable to capture the high-level semantic anatomical dense correspondences. In this work, we propose a novel multi-task learning system, JSSR, based on an end-to-end 3D convolutional neural network that is composed of a generator, a register and a segmentor, for the tasks of synthesis, registration and segmentation, respectively. This system is optimized to satisfy the implicit constraints between different tasks unsupervisedly. It first synthesizes the source domain images into the target domain, then an intra-modal registration is applied on the synthesized images and target images. Then we can get the semantic segmentation by applying segmentors on the synthesized images and target images, which are aligned by the same deformation field generated by the registers. The supervision from another fully-annotated dataset is used to regularize the segmentors. We extensively evaluate our JSSR system on a large-scale medical image dataset containing 1,485 patient CT imaging studies of four different phases (i.e., 5,940 3D CT scans with pathological livers) on the registration, segmentation and synthesis tasks. The performance is improved after joint training on the registration and segmentation tasks by 0.9% and 1.9% respectively from a highly competitive and accurate baseline. The registration part also consistently outperforms the conventional state-of-the-art multi-modal registration methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge