JinHyeong Park
Inpainting Pathology in Lumbar Spine MRI with Latent Diffusion
Jun 04, 2024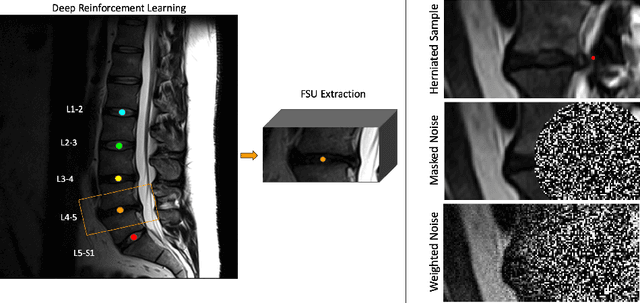
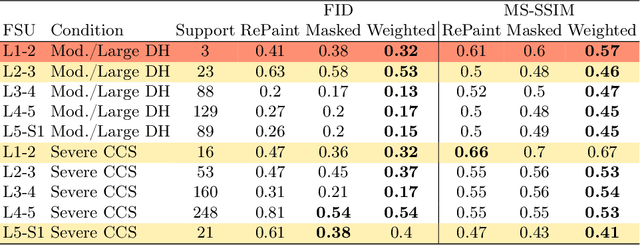
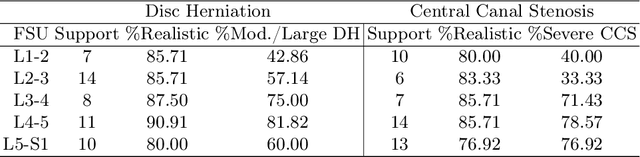
Abstract:Data driven models for automated diagnosis in radiology suffer from insufficient and imbalanced datasets due to low representation of pathology in a population and the cost of expert annotations. Datasets can be bolstered through data augmentation. However, even when utilizing a full suite of transformations during model training, typical data augmentations do not address variations in human anatomy. An alternative direction is to synthesize data using generative models, which can potentially craft datasets with specific attributes. While this holds promise, commonly used generative models such as Generative Adversarial Networks may inadvertently produce anatomically inaccurate features. On the other hand, diffusion models, which offer greater stability, tend to memorize training data, raising concerns about privacy and generative diversity. Alternatively, inpainting has the potential to augment data through directly inserting pathology in medical images. However, this approach introduces a new challenge: accurately merging the generated pathological features with the surrounding anatomical context. While inpainting is a well established method for addressing simple lesions, its application to pathologies that involve complex structural changes remains relatively unexplored. We propose an efficient method for inpainting pathological features onto healthy anatomy in MRI through voxelwise noise scheduling in a latent diffusion model. We evaluate the method's ability to insert disc herniation and central canal stenosis in lumbar spine sagittal T2 MRI, and it achieves superior Frechet Inception Distance compared to state-of-the-art methods.
Less is More: Simultaneous View Classification and Landmark Detection for Abdominal Ultrasound Images
Jun 04, 2018



Abstract:An abdominal ultrasound examination, which is the most common ultrasound examination, requires substantial manual efforts to acquire standard abdominal organ views, annotate the views in texts, and record clinically relevant organ measurements. Hence, automatic view classification and landmark detection of the organs can be instrumental to streamline the examination workflow. However, this is a challenging problem given not only the inherent difficulties from the ultrasound modality, e.g., low contrast and large variations, but also the heterogeneity across tasks, i.e., one classification task for all views, and then one landmark detection task for each relevant view. While convolutional neural networks (CNN) have demonstrated more promising outcomes on ultrasound image analytics than traditional machine learning approaches, it becomes impractical to deploy multiple networks (one for each task) due to the limited computational and memory resources on most existing ultrasound scanners. To overcome such limits, we propose a multi-task learning framework to handle all the tasks by a single network. This network is integrated to perform view classification and landmark detection simultaneously; it is also equipped with global convolutional kernels, coordinate constraints, and a conditional adversarial module to leverage the performances. In an experimental study based on 187,219 ultrasound images, with the proposed simplified approach we achieve (1) view classification accuracy better than the agreement between two clinical experts and (2) landmark-based measurement errors on par with inter-user variability. The multi-task approach also benefits from sharing the feature extraction during the training process across all tasks and, as a result, outperforms the approaches that address each task individually.
Automatic Vertebra Labeling in Large-Scale 3D CT using Deep Image-to-Image Network with Message Passing and Sparsity Regularization
May 17, 2017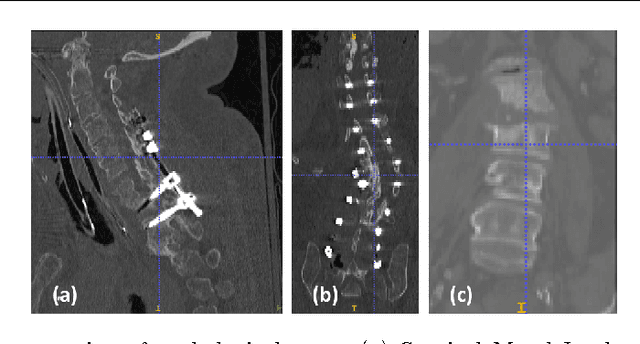
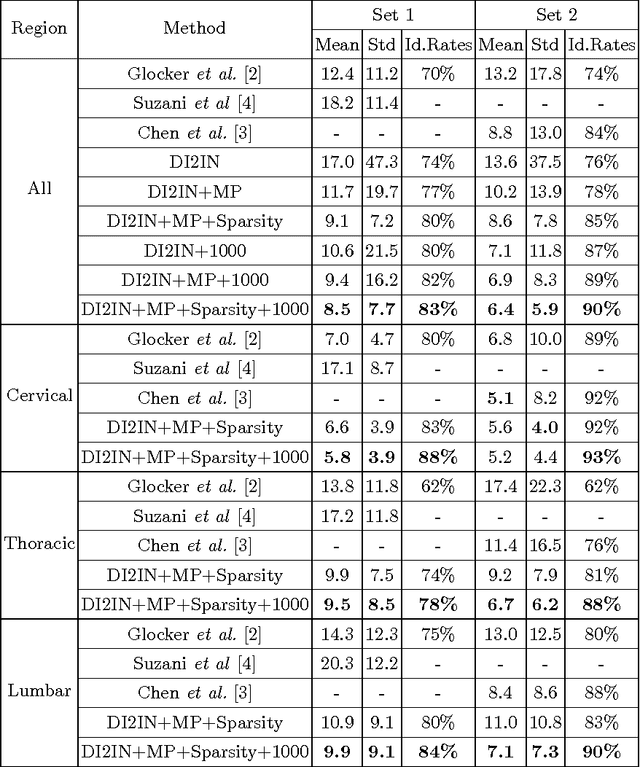
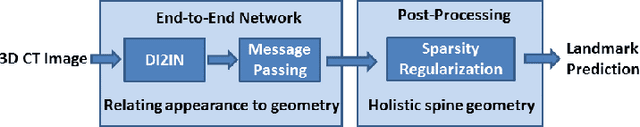
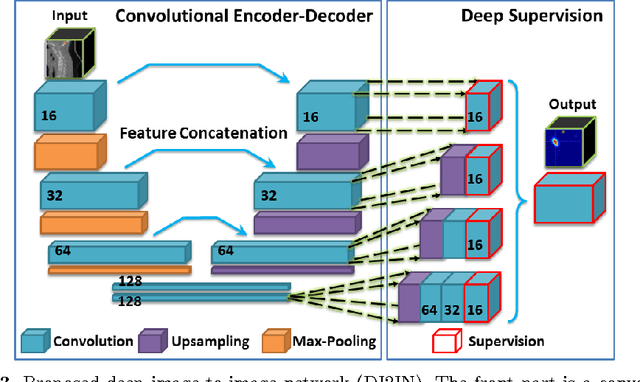
Abstract:Automatic localization and labeling of vertebra in 3D medical images plays an important role in many clinical tasks, including pathological diagnosis, surgical planning and postoperative assessment. However, the unusual conditions of pathological cases, such as the abnormal spine curvature, bright visual imaging artifacts caused by metal implants, and the limited field of view, increase the difficulties of accurate localization. In this paper, we propose an automatic and fast algorithm to localize and label the vertebra centroids in 3D CT volumes. First, we deploy a deep image-to-image network (DI2IN) to initialize vertebra locations, employing the convolutional encoder-decoder architecture together with multi-level feature concatenation and deep supervision. Next, the centroid probability maps from DI2IN are iteratively evolved with the message passing schemes based on the mutual relation of vertebra centroids. Finally, the localization results are refined with sparsity regularization. The proposed method is evaluated on a public dataset of 302 spine CT volumes with various pathologies. Our method outperforms other state-of-the-art methods in terms of localization accuracy. The run time is around 3 seconds on average per case. To further boost the performance, we retrain the DI2IN on additional 1000+ 3D CT volumes from different patients. To the best of our knowledge, this is the first time more than 1000 3D CT volumes with expert annotation are adopted in experiments for the anatomic landmark detection tasks. Our experimental results show that training with such a large dataset significantly improves the performance and the overall identification rate, for the first time by our knowledge, reaches 90 %.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge