Shun Miao
Lumbar Bone Mineral Density Estimation from Chest X-ray Images: Anatomy-aware Attentive Multi-ROI Modeling
Jan 05, 2022



Abstract:Osteoporosis is a common chronic metabolic bone disease that is often under-diagnosed and under-treated due to the limited access to bone mineral density (BMD) examinations, e.g. via Dual-energy X-ray Absorptiometry (DXA). In this paper, we propose a method to predict BMD from Chest X-ray (CXR), one of the most commonly accessible and low-cost medical imaging examinations. Our method first automatically detects Regions of Interest (ROIs) of local and global bone structures from the CXR. Then a multi-ROI deep model with transformer encoder is developed to exploit both local and global information in the chest X-ray image for accurate BMD estimation. Our method is evaluated on 13719 CXR patient cases with their ground truth BMD scores measured by gold-standard DXA. The model predicted BMD has a strong correlation with the ground truth (Pearson correlation coefficient 0.889 on lumbar 1). When applied for osteoporosis screening, it achieves a high classification performance (AUC 0.963 on lumbar 1). As the first effort in the field using CXR scans to predict the BMD, the proposed algorithm holds strong potential in early osteoporosis screening and public health promotion.
Coherence Learning using Keypoint-based Pooling Network for Accurately Assessing Radiographic Knee Osteoarthritis
Dec 16, 2021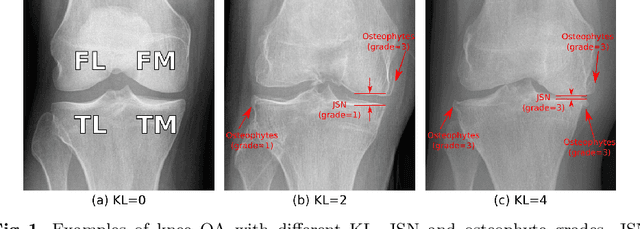
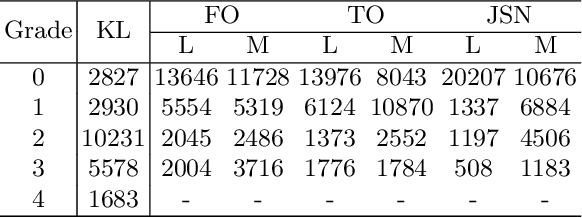
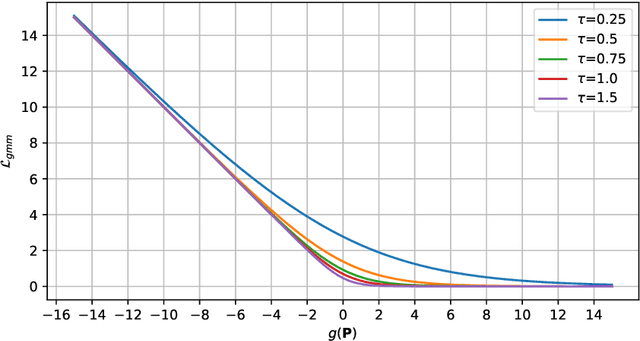
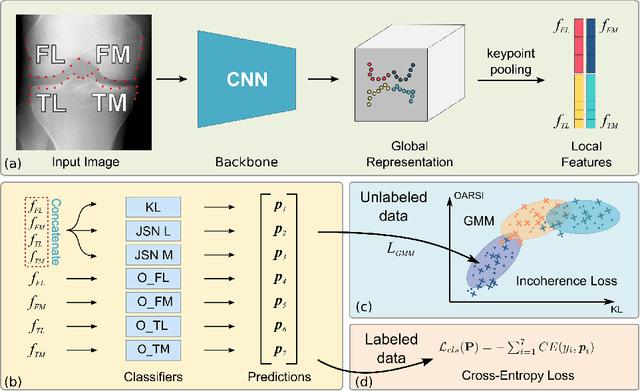
Abstract:Knee osteoarthritis (OA) is a common degenerate joint disorder that affects a large population of elderly people worldwide. Accurate radiographic assessment of knee OA severity plays a critical role in chronic patient management. Current clinically-adopted knee OA grading systems are observer subjective and suffer from inter-rater disagreements. In this work, we propose a computer-aided diagnosis approach to provide more accurate and consistent assessments of both composite and fine-grained OA grades simultaneously. A novel semi-supervised learning method is presented to exploit the underlying coherence in the composite and fine-grained OA grades by learning from unlabeled data. By representing the grade coherence using the log-probability of a pre-trained Gaussian Mixture Model, we formulate an incoherence loss to incorporate unlabeled data in training. The proposed method also describes a keypoint-based pooling network, where deep image features are pooled from the disease-targeted keypoints (extracted along the knee joint) to provide more aligned and pathologically informative feature representations, for accurate OA grade assessments. The proposed method is comprehensively evaluated on the public Osteoarthritis Initiative (OAI) data, a multi-center ten-year observational study on 4,796 subjects. Experimental results demonstrate that our method leads to significant improvements over previous strong whole image-based deep classification network baselines (like ResNet-50).
Scalable Semi-supervised Landmark Localization for X-ray Images using Few-shot Deep Adaptive Graph
Apr 29, 2021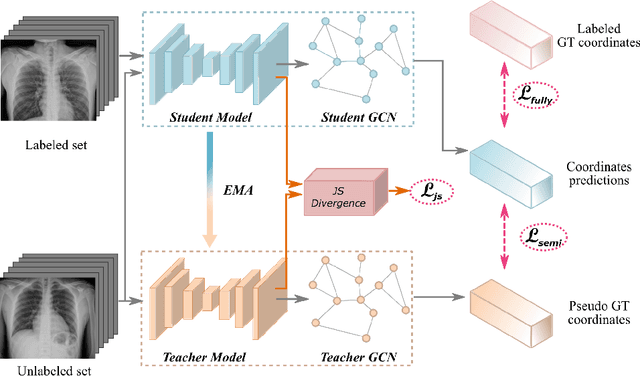


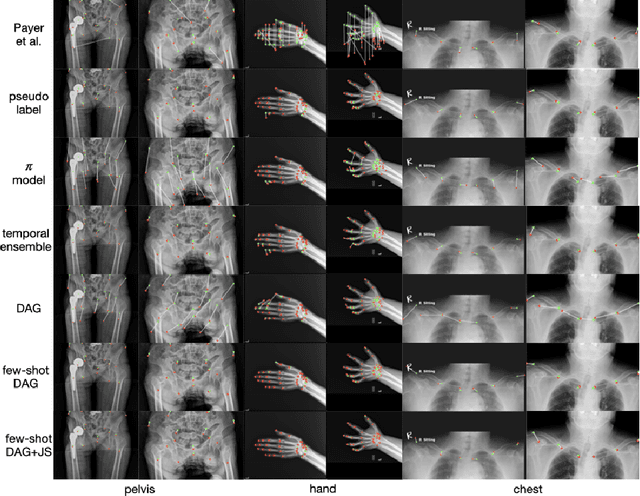
Abstract:Landmark localization plays an important role in medical image analysis. Learning based methods, including CNN and GCN, have demonstrated the state-of-the-art performance. However, most of these methods are fully-supervised and heavily rely on manual labeling of a large training dataset. In this paper, based on a fully-supervised graph-based method, DAG, we proposed a semi-supervised extension of it, termed few-shot DAG, \ie five-shot DAG. It first trains a DAG model on the labeled data and then fine-tunes the pre-trained model on the unlabeled data with a teacher-student SSL mechanism. In addition to the semi-supervised loss, we propose another loss using JS divergence to regulate the consistency of the intermediate feature maps. We extensively evaluated our method on pelvis, hand and chest landmark detection tasks. Our experiment results demonstrate consistent and significant improvements over previous methods.
Deep Implicit Statistical Shape Models for 3D Medical Image Delineation
Apr 07, 2021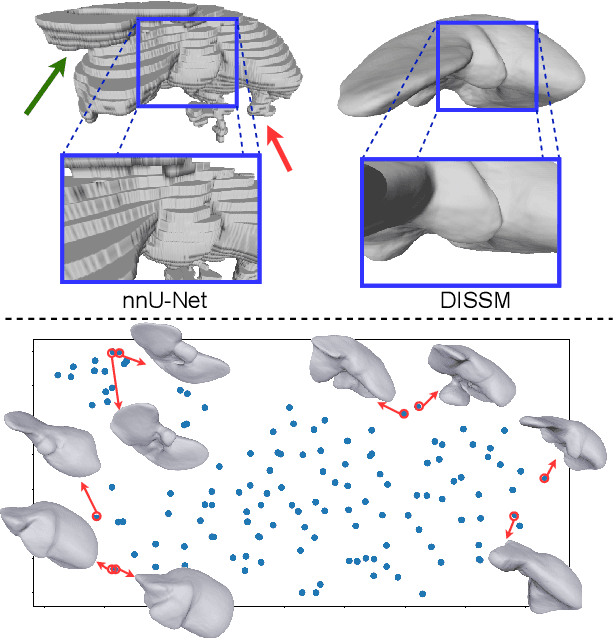
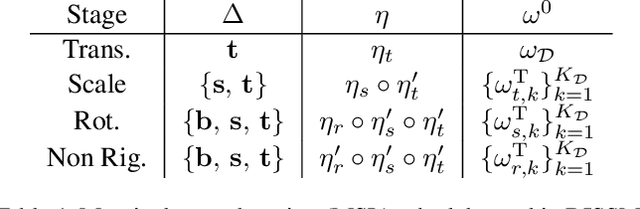
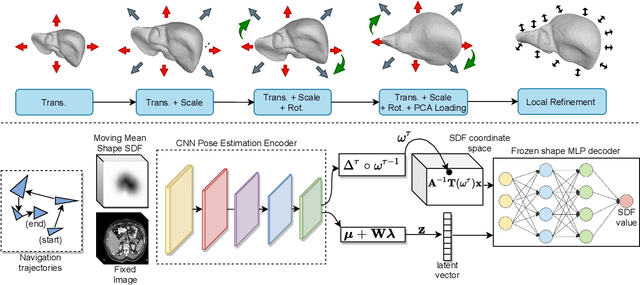
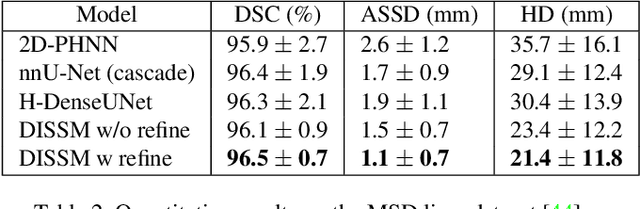
Abstract:3D delineation of anatomical structures is a cardinal goal in medical imaging analysis. Prior to deep learning, statistical shape models that imposed anatomical constraints and produced high quality surfaces were a core technology. Prior to deep learning, statistical shape models that imposed anatomical constraints and produced high quality surfaces were a core technology. Today fully-convolutional networks (FCNs), while dominant, do not offer these capabilities. We present deep implicit statistical shape models (DISSMs), a new approach to delineation that marries the representation power of convolutional neural networks (CNNs) with the robustness of SSMs. DISSMs use a deep implicit surface representation to produce a compact and descriptive shape latent space that permits statistical models of anatomical variance. To reliably fit anatomically plausible shapes to an image, we introduce a novel rigid and non-rigid pose estimation pipeline that is modelled as a Markov decision process(MDP). We outline a training regime that includes inverted episodic training and a deep realization of marginal space learning (MSL). Intra-dataset experiments on the task of pathological liver segmentation demonstrate that DISSMs can perform more robustly than three leading FCN models, including nnU-Net: reducing the mean Hausdorff distance (HD) by 7.7-14.3mm and improving the worst case Dice-Sorensen coefficient (DSC) by 1.2-2.3%. More critically, cross-dataset experiments on a dataset directly reflecting clinical deployment scenarios demonstrate that DISSMs improve the mean DSC and HD by 3.5-5.9% and 12.3-24.5mm, respectively, and the worst-case DSC by 5.4-7.3%. These improvements are over and above any benefits from representing delineations with high-quality surface.
Opportunistic Screening of Osteoporosis Using Plain Film Chest X-ray
Apr 05, 2021



Abstract:Osteoporosis is a common chronic metabolic bone disease that is often under-diagnosed and under-treated due to the limited access to bone mineral density (BMD) examinations, Dual-energy X-ray Absorptiometry (DXA). In this paper, we propose a method to predict BMD from Chest X-ray (CXR), one of the most common, accessible, and low-cost medical image examinations. Our method first automatically detects Regions of Interest (ROIs) of local and global bone structures from the CXR. Then a multi-ROI model is developed to exploit both local and global information in the chest X-ray image for accurate BMD estimation. Our method is evaluated on 329 CXR cases with ground truth BMD measured by DXA. The model predicted BMD has a strong correlation with the gold standard DXA BMD (Pearson correlation coefficient 0.840). When applied for osteoporosis screening, it achieves a high classification performance (AUC 0.936). As the first effort in the field to use CXR scans to predict the spine BMD, the proposed algorithm holds strong potential in enabling early osteoporosis screening through routine chest X-rays and contributing to the enhancement of public health.
Semi-Supervised Learning for Bone Mineral Density Estimation in Hip X-ray Images
Mar 24, 2021



Abstract:Bone mineral density (BMD) is a clinically critical indicator of osteoporosis, usually measured by dual-energy X-ray absorptiometry (DEXA). Due to the limited accessibility of DEXA machines and examinations, osteoporosis is often under-diagnosed and under-treated, leading to increased fragility fracture risks. Thus it is highly desirable to obtain BMDs with alternative cost-effective and more accessible medical imaging examinations such as X-ray plain films. In this work, we formulate the BMD estimation from plain hip X-ray images as a regression problem. Specifically, we propose a new semi-supervised self-training algorithm to train the BMD regression model using images coupled with DEXA measured BMDs and unlabeled images with pseudo BMDs. Pseudo BMDs are generated and refined iteratively for unlabeled images during self-training. We also present a novel adaptive triplet loss to improve the model's regression accuracy. On an in-house dataset of 1,090 images (819 unique patients), our BMD estimation method achieves a high Pearson correlation coefficient of 0.8805 to ground-truth BMDs. It offers good feasibility to use the more accessible and cheaper X-ray imaging for opportunistic osteoporosis screening.
A New Window Loss Function for Bone Fracture Detection and Localization in X-ray Images with Point-based Annotation
Jan 04, 2021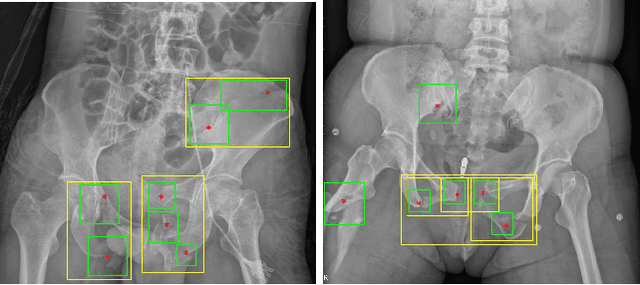
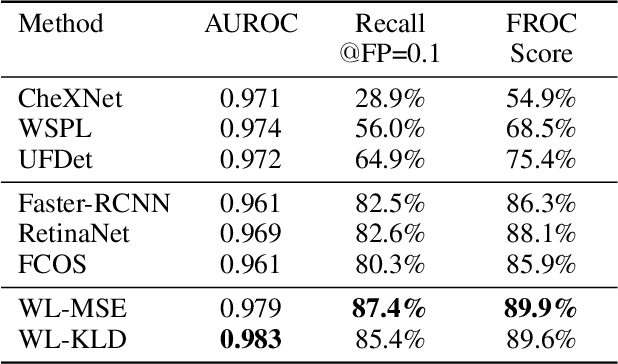
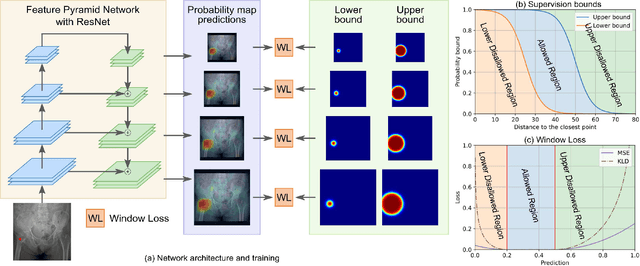
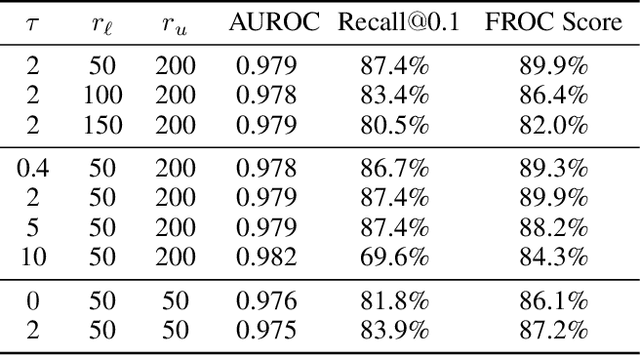
Abstract:Object detection methods are widely adopted for computer-aided diagnosis using medical images. Anomalous findings are usually treated as objects that are described by bounding boxes. Yet, many pathological findings, e.g., bone fractures, cannot be clearly defined by bounding boxes, owing to considerable instance, shape and boundary ambiguities. This makes bounding box annotations, and their associated losses, highly ill-suited. In this work, we propose a new bone fracture detection method for X-ray images, based on a labor effective and flexible annotation scheme suitable for abnormal findings with no clear object-level spatial extents or boundaries. Our method employs a simple, intuitive, and informative point-based annotation protocol to mark localized pathology information. To address the uncertainty in the fracture scales annotated via point(s), we convert the annotations into pixel-wise supervision that uses lower and upper bounds with positive, negative, and uncertain regions. A novel Window Loss is subsequently proposed to only penalize the predictions outside of the uncertain regions. Our method has been extensively evaluated on 4410 pelvic X-ray images of unique patients. Experiments demonstrate that our method outperforms previous state-of-the-art image classification and object detection baselines by healthy margins, with an AUROC of 0.983 and FROC score of 89.6%.
Knowledge Distillation with Adaptive Asymmetric Label Sharpening for Semi-supervised Fracture Detection in Chest X-rays
Dec 30, 2020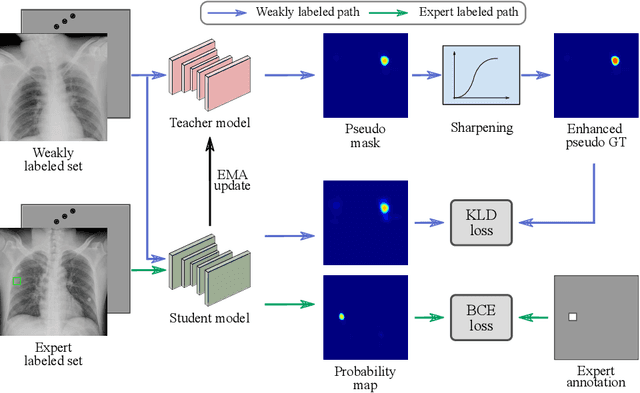
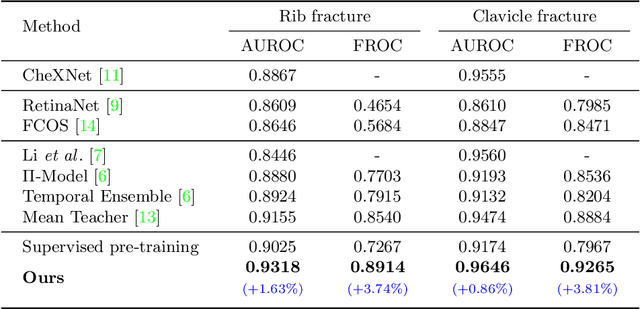
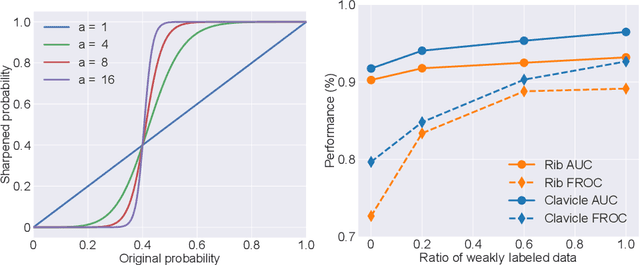
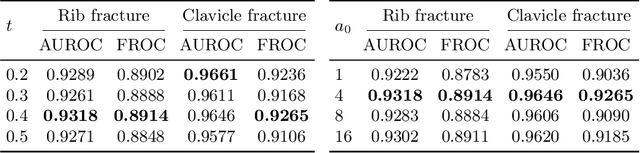
Abstract:Exploiting available medical records to train high performance computer-aided diagnosis (CAD) models via the semi-supervised learning (SSL) setting is emerging to tackle the prohibitively high labor costs involved in large-scale medical image annotations. Despite the extensive attentions received on SSL, previous methods failed to 1) account for the low disease prevalence in medical records and 2) utilize the image-level diagnosis indicated from the medical records. Both issues are unique to SSL for CAD models. In this work, we propose a new knowledge distillation method that effectively exploits large-scale image-level labels extracted from the medical records, augmented with limited expert annotated region-level labels, to train a rib and clavicle fracture CAD model for chest X-ray (CXR). Our method leverages the teacher-student model paradigm and features a novel adaptive asymmetric label sharpening (AALS) algorithm to address the label imbalance problem that specially exists in medical domain. Our approach is extensively evaluated on all CXR (N = 65,845) from the trauma registry of anonymous hospital over a period of 9 years (2008-2016), on the most common rib and clavicle fractures. The experiment results demonstrate that our method achieves the state-of-the-art fracture detection performance, i.e., an area under receiver operating characteristic curve (AUROC) of 0.9318 and a free-response receiver operating characteristic (FROC) score of 0.8914 on the rib fractures, significantly outperforming previous approaches by an AUROC gap of 1.63% and an FROC improvement by 3.74%. Consistent performance gains are also observed for clavicle fracture detection.
Automatic Vertebra Localization and Identification in CT by Spine Rectification and Anatomically-constrained Optimization
Dec 14, 2020



Abstract:Accurate vertebra localization and identification are required in many clinical applications of spine disorder diagnosis and surgery planning. However, significant challenges are posed in this task by highly varying pathologies (such as vertebral compression fracture, scoliosis, and vertebral fixation) and imaging conditions (such as limited field of view and metal streak artifacts). This paper proposes a robust and accurate method that effectively exploits the anatomical knowledge of the spine to facilitate vertebra localization and identification. A key point localization model is trained to produce activation maps of vertebra centers. They are then re-sampled along the spine centerline to produce spine-rectified activation maps, which are further aggregated into 1-D activation signals. Following this, an anatomically-constrained optimization module is introduced to jointly search for the optimal vertebra centers under a soft constraint that regulates the distance between vertebrae and a hard constraint on the consecutive vertebra indices. When being evaluated on a major public benchmark of 302 highly pathological CT images, the proposed method reports the state of the art identification (id.) rate of 97.4%, and outperforms the best competing method of 94.7% id. rate by reducing the relative id. error rate by half.
Self-supervised Learning of Pixel-wise Anatomical Embeddings in Radiological Images
Dec 04, 2020



Abstract:Radiological images such as computed tomography (CT) and X-rays render anatomy with intrinsic structures. Being able to reliably locate the same anatomical or semantic structure across varying images is a fundamental task in medical image analysis. In principle it is possible to use landmark detection or semantic segmentation for this task, but to work well these require large numbers of labeled data for each anatomical structure and sub-structure of interest. A more universal approach would discover the intrinsic structure from unlabeled images. We introduce such an approach, called Self-supervised Anatomical eMbedding (SAM). SAM generates semantic embeddings for each image pixel that describes its anatomical location or body part. To produce such embeddings, we propose a pixel-level contrastive learning framework. A coarse-to-fine strategy ensures both global and local anatomical information are encoded. Negative sample selection strategies are designed to enhance the discriminability among different body parts. Using SAM, one can label any point of interest on a template image, and then locate the same body part in other images by simple nearest neighbor searching. We demonstrate the effectiveness of SAM in multiple tasks with 2D and 3D image modalities. On a chest CT dataset with 19 landmarks, SAM outperforms widely-used registration algorithms while being 200 times faster. On two X-ray datasets, SAM, with only one labeled template image, outperforms supervised methods trained on 50 labeled images. We also apply SAM on whole-body follow-up lesion matching in CT and obtain an accuracy of 91%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge