Weijian Li
Adaptive Batch Size Schedules for Distributed Training of Language Models with Data and Model Parallelism
Dec 30, 2024
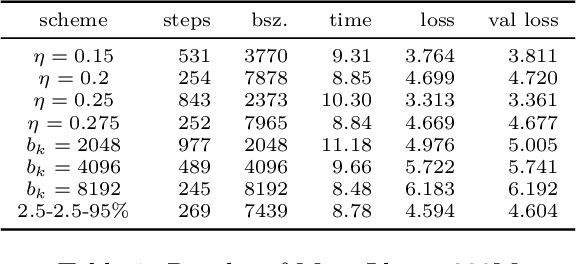

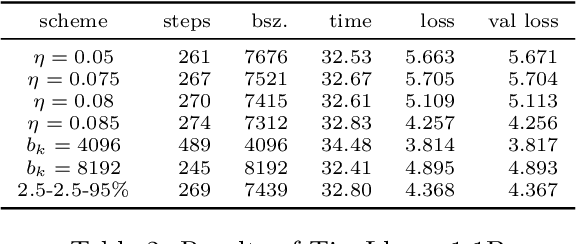
Abstract:An appropriate choice of batch sizes in large-scale model training is crucial, yet it involves an intrinsic yet inevitable dilemma: large-batch training improves training efficiency in terms of memory utilization, while generalization performance often deteriorates due to small amounts of gradient noise. Despite this dilemma, the common practice of choosing batch sizes in language model training often prioritizes training efficiency -- employing either constant large sizes with data parallelism or implementing batch size warmup schedules. However, such batch size schedule designs remain heuristic and often fail to adapt to training dynamics, presenting the challenge of designing adaptive batch size schedules. Given the abundance of available datasets and the data-hungry nature of language models, data parallelism has become an indispensable distributed training paradigm, enabling the use of larger batch sizes for gradient computation. However, vanilla data parallelism requires replicas of model parameters, gradients, and optimizer states at each worker, which prohibits training larger models with billions of parameters. To optimize memory usage, more advanced parallelism strategies must be employed. In this work, we propose general-purpose and theoretically principled adaptive batch size schedules compatible with data parallelism and model parallelism. We develop a practical implementation with PyTorch Fully Sharded Data Parallel, facilitating the pretraining of language models of different sizes. We empirically demonstrate that our proposed approaches outperform constant batch sizes and heuristic batch size warmup schedules in the pretraining of models in the Llama family, with particular focus on smaller models with up to 3 billion parameters. We also establish theoretical convergence guarantees for such adaptive batch size schedules with Adam for general smooth nonconvex objectives.
CRTRE: Causal Rule Generation with Target Trial Emulation Framework
Nov 10, 2024

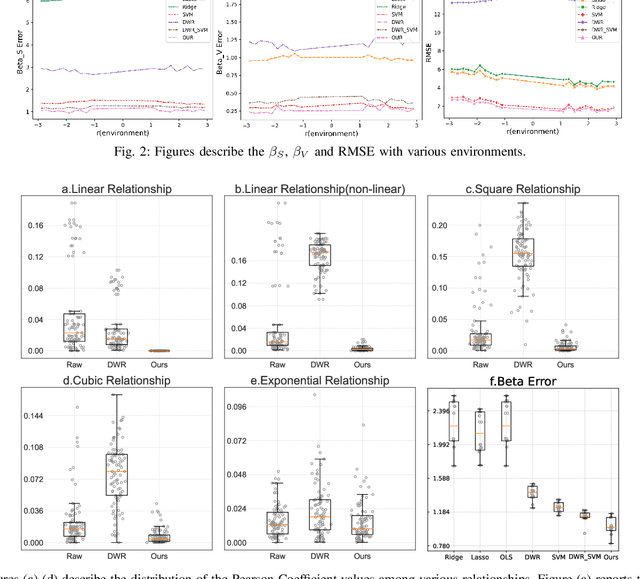

Abstract:Causal inference and model interpretability are gaining increasing attention, particularly in the biomedical domain. Despite recent advance, decorrelating features in nonlinear environments with human-interpretable representations remains underexplored. In this study, we introduce a novel method called causal rule generation with target trial emulation framework (CRTRE), which applies randomize trial design principles to estimate the causal effect of association rules. We then incorporate such association rules for the downstream applications such as prediction of disease onsets. Extensive experiments on six healthcare datasets, including synthetic data, real-world disease collections, and MIMIC-III/IV, demonstrate the model's superior performance. Specifically, our method achieved a $\beta$ error of 0.907, outperforming DWR (1.024) and SVM (1.141). On real-world datasets, our model achieved accuracies of 0.789, 0.920, and 0.300 for Esophageal Cancer, Heart Disease, and Cauda Equina Syndrome prediction task, respectively, consistently surpassing baseline models. On the ICD code prediction tasks, it achieved AUC Macro scores of 92.8 on MIMIC-III and 96.7 on MIMIC-IV, outperforming the state-of-the-art models KEPT and MSMN. Expert evaluations further validate the model's effectiveness, causality, and interpretability.
Communication-Efficient Adaptive Batch Size Strategies for Distributed Local Gradient Methods
Jun 20, 2024



Abstract:Modern deep neural networks often require distributed training with many workers due to their large size. As worker numbers increase, communication overheads become the main bottleneck in data-parallel minibatch stochastic gradient methods with per-iteration gradient synchronization. Local gradient methods like Local SGD reduce communication by only syncing after several local steps. Despite understanding their convergence in i.i.d. and heterogeneous settings and knowing the importance of batch sizes for efficiency and generalization, optimal local batch sizes are difficult to determine. We introduce adaptive batch size strategies for local gradient methods that increase batch sizes adaptively to reduce minibatch gradient variance. We provide convergence guarantees under homogeneous data conditions and support our claims with image classification experiments, demonstrating the effectiveness of our strategies in training and generalization.
BiSHop: Bi-Directional Cellular Learning for Tabular Data with Generalized Sparse Modern Hopfield Model
Apr 04, 2024



Abstract:We introduce the \textbf{B}i-Directional \textbf{S}parse \textbf{Hop}field Network (\textbf{BiSHop}), a novel end-to-end framework for deep tabular learning. BiSHop handles the two major challenges of deep tabular learning: non-rotationally invariant data structure and feature sparsity in tabular data. Our key motivation comes from the recent established connection between associative memory and attention mechanisms. Consequently, BiSHop uses a dual-component approach, sequentially processing data both column-wise and row-wise through two interconnected directional learning modules. Computationally, these modules house layers of generalized sparse modern Hopfield layers, a sparse extension of the modern Hopfield model with adaptable sparsity. Methodologically, BiSHop facilitates multi-scale representation learning, capturing both intra-feature and inter-feature interactions, with adaptive sparsity at each scale. Empirically, through experiments on diverse real-world datasets, we demonstrate that BiSHop surpasses current SOTA methods with significantly less HPO runs, marking it a robust solution for deep tabular learning.
Outlier-Efficient Hopfield Layers for Large Transformer-Based Models
Apr 04, 2024



Abstract:We introduce an Outlier-Efficient Modern Hopfield Model (termed $\mathtt{OutEffHop}$) and use it to address the outlier-induced challenge of quantizing gigantic transformer-based models. Our main contribution is a novel associative memory model facilitating \textit{outlier-efficient} associative memory retrievals. Interestingly, this memory model manifests a model-based interpretation of an outlier-efficient attention mechanism ($\text{Softmax}_1$): it is an approximation of the memory retrieval process of $\mathtt{OutEffHop}$. Methodologically, this allows us to debut novel outlier-efficient Hopfield layers a powerful attention alternative with superior post-quantization performance. Theoretically, the Outlier-Efficient Modern Hopfield Model retains and improves the desirable properties of the standard modern Hopfield models, including fixed point convergence and exponential storage capacity. Empirically, we demonstrate the proposed model's efficacy across large-scale transformer-based and Hopfield-based models (including BERT, OPT, ViT and STanHop-Net), benchmarking against state-of-the-art methods including $\mathtt{Clipped\_Softmax}$ and $\mathtt{Gated\_Attention}$. Notably, $\mathtt{OutEffHop}$ achieves on average $\sim$22+\% reductions in both average kurtosis and maximum infinity norm of model outputs accross 4 models.
STanHop: Sparse Tandem Hopfield Model for Memory-Enhanced Time Series Prediction
Dec 28, 2023



Abstract:We present STanHop-Net (Sparse Tandem Hopfield Network) for multivariate time series prediction with memory-enhanced capabilities. At the heart of our approach is STanHop, a novel Hopfield-based neural network block, which sparsely learns and stores both temporal and cross-series representations in a data-dependent fashion. In essence, STanHop sequentially learn temporal representation and cross-series representation using two tandem sparse Hopfield layers. In addition, StanHop incorporates two additional external memory modules: a Plug-and-Play module and a Tune-and-Play module for train-less and task-aware memory-enhancements, respectively. They allow StanHop-Net to swiftly respond to certain sudden events. Methodologically, we construct the StanHop-Net by stacking STanHop blocks in a hierarchical fashion, enabling multi-resolution feature extraction with resolution-specific sparsity. Theoretically, we introduce a sparse extension of the modern Hopfield model (Generalized Sparse Modern Hopfield Model) and show that it endows a tighter memory retrieval error compared to the dense counterpart without sacrificing memory capacity. Empirically, we validate the efficacy of our framework on both synthetic and real-world settings.
DocTr: Document Transformer for Structured Information Extraction in Documents
Jul 16, 2023Abstract:We present a new formulation for structured information extraction (SIE) from visually rich documents. It aims to address the limitations of existing IOB tagging or graph-based formulations, which are either overly reliant on the correct ordering of input text or struggle with decoding a complex graph. Instead, motivated by anchor-based object detectors in vision, we represent an entity as an anchor word and a bounding box, and represent entity linking as the association between anchor words. This is more robust to text ordering, and maintains a compact graph for entity linking. The formulation motivates us to introduce 1) a DOCument TRansformer (DocTr) that aims at detecting and associating entity bounding boxes in visually rich documents, and 2) a simple pre-training strategy that helps learn entity detection in the context of language. Evaluations on three SIE benchmarks show the effectiveness of the proposed formulation, and the overall approach outperforms existing solutions.
DNABERT-2: Efficient Foundation Model and Benchmark For Multi-Species Genome
Jun 26, 2023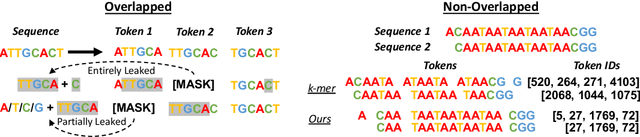
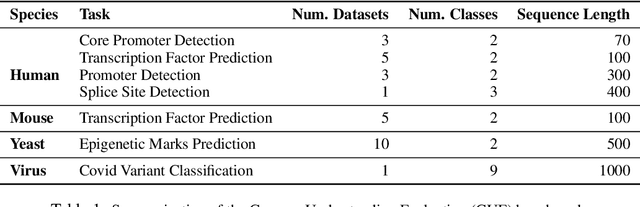
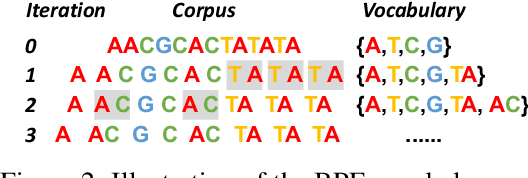
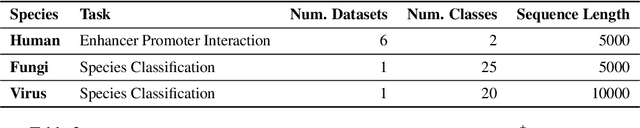
Abstract:Decoding the linguistic intricacies of the genome is a crucial problem in biology, and pre-trained foundational models such as DNABERT and Nucleotide Transformer have made significant strides in this area. Existing works have largely hinged on k-mer, fixed-length permutations of A, T, C, and G, as the token of the genome language due to its simplicity. However, we argue that the computation and sample inefficiencies introduced by k-mer tokenization are primary obstacles in developing large genome foundational models. We provide conceptual and empirical insights into genome tokenization, building on which we propose to replace k-mer tokenization with Byte Pair Encoding (BPE), a statistics-based data compression algorithm that constructs tokens by iteratively merging the most frequent co-occurring genome segment in the corpus. We demonstrate that BPE not only overcomes the limitations of k-mer tokenization but also benefits from the computational efficiency of non-overlapping tokenization. Based on these insights, we introduce DNABERT-2, a refined genome foundation model that adapts an efficient tokenizer and employs multiple strategies to overcome input length constraints, reduce time and memory expenditure, and enhance model capability. Furthermore, we identify the absence of a comprehensive and standardized benchmark for genome understanding as another significant impediment to fair comparative analysis. In response, we propose the Genome Understanding Evaluation (GUE), a comprehensive multi-species genome classification dataset that amalgamates $28$ distinct datasets across $7$ tasks, with input lengths ranging from $70$ to $1000$. Through comprehensive experiments on the GUE benchmark, we demonstrate that DNABERT-2 achieves comparable performance to the state-of-the-art model with $21 \times$ fewer parameters and approximately $56 \times$ less GPU time in pre-training.
Feature Programming for Multivariate Time Series Prediction
Jun 09, 2023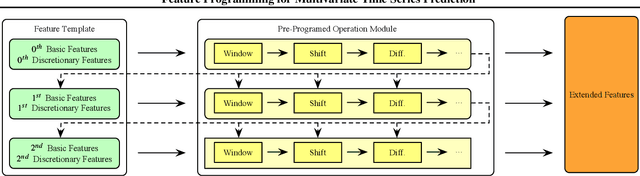
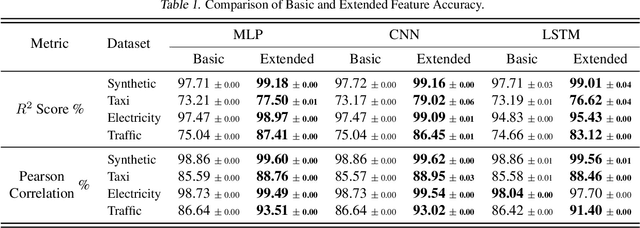
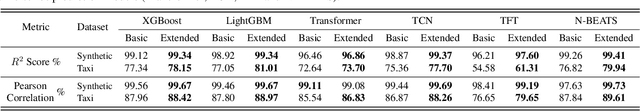
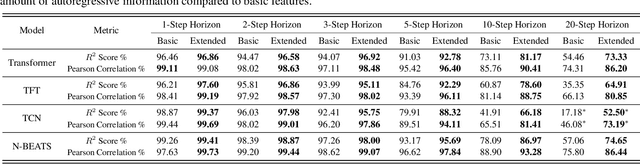
Abstract:We introduce the concept of programmable feature engineering for time series modeling and propose a feature programming framework. This framework generates large amounts of predictive features for noisy multivariate time series while allowing users to incorporate their inductive bias with minimal effort. The key motivation of our framework is to view any multivariate time series as a cumulative sum of fine-grained trajectory increments, with each increment governed by a novel spin-gas dynamical Ising model. This fine-grained perspective motivates the development of a parsimonious set of operators that summarize multivariate time series in an abstract fashion, serving as the foundation for large-scale automated feature engineering. Numerically, we validate the efficacy of our method on several synthetic and real-world noisy time series datasets.
Unsupervised Self-Driving Attention Prediction via Uncertainty Mining and Knowledge Embedding
Mar 22, 2023



Abstract:Predicting attention regions of interest is an important yet challenging task for self-driving systems. Existing methodologies rely on large-scale labeled traffic datasets that are labor-intensive to obtain. Besides, the huge domain gap between natural scenes and traffic scenes in current datasets also limits the potential for model training. To address these challenges, we are the first to introduce an unsupervised way to predict self-driving attention by uncertainty modeling and driving knowledge integration. Our approach's Uncertainty Mining Branch (UMB) discovers commonalities and differences from multiple generated pseudo-labels achieved from models pre-trained on natural scenes by actively measuring the uncertainty. Meanwhile, our Knowledge Embedding Block (KEB) bridges the domain gap by incorporating driving knowledge to adaptively refine the generated pseudo-labels. Quantitative and qualitative results with equivalent or even more impressive performance compared to fully-supervised state-of-the-art approaches across all three public datasets demonstrate the effectiveness of the proposed method and the potential of this direction. The code will be made publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge