Zhihan Zhou
AncientBench: Towards Comprehensive Evaluation on Excavated and Transmitted Chinese Corpora
Dec 19, 2025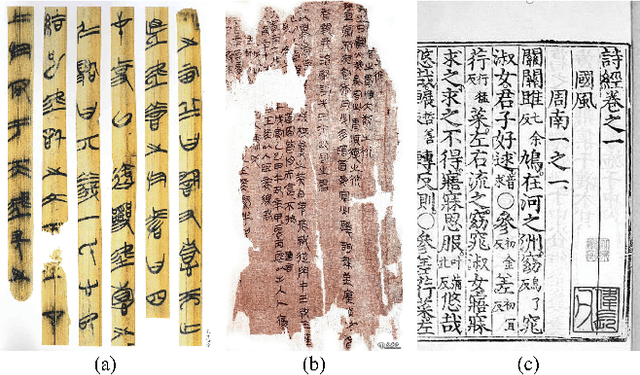
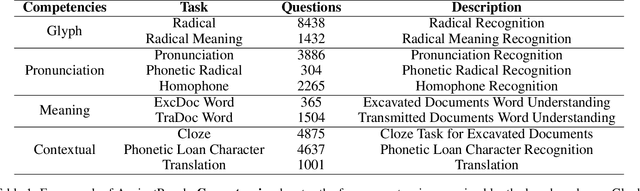

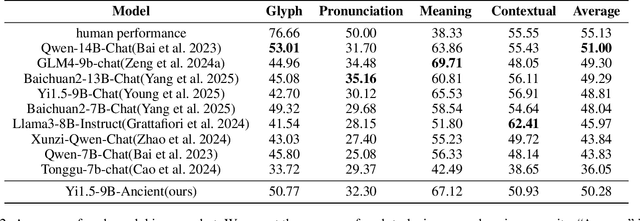
Abstract:Comprehension of ancient texts plays an important role in archaeology and understanding of Chinese history and civilization. The rapid development of large language models needs benchmarks that can evaluate their comprehension of ancient characters. Existing Chinese benchmarks are mostly targeted at modern Chinese and transmitted documents in ancient Chinese, but the part of excavated documents in ancient Chinese is not covered. To meet this need, we propose the AncientBench, which aims to evaluate the comprehension of ancient characters, especially in the scenario of excavated documents. The AncientBench is divided into four dimensions, which correspond to the four competencies of ancient character comprehension: glyph comprehension, pronunciation comprehension, meaning comprehension, and contextual comprehension. The benchmark also contains ten tasks, including radical, phonetic radical, homophone, cloze, translation, and more, providing a comprehensive framework for evaluation. We convened archaeological researchers to conduct experimental evaluations, proposed an ancient model as baseline, and conducted extensive experiments on the currently best-performing large language models. The experimental results reveal the great potential of large language models in ancient textual scenarios as well as the gap with humans. Our research aims to promote the development and application of large language models in the field of archaeology and ancient Chinese language.
DeepVRegulome: DNABERT-based deep-learning framework for predicting the functional impact of short genomic variants on the human regulome
Nov 12, 2025



Abstract:Whole-genome sequencing (WGS) has revealed numerous non-coding short variants whose functional impacts remain poorly understood. Despite recent advances in deep-learning genomic approaches, accurately predicting and prioritizing clinically relevant mutations in gene regulatory regions remains a major challenge. Here we introduce Deep VRegulome, a deep-learning method for prediction and interpretation of functionally disruptive variants in the human regulome, which combines 700 DNABERT fine-tuned models, trained on vast amounts of ENCODE gene regulatory regions, with variant scoring, motif analysis, attention-based visualization, and survival analysis. We showcase its application on TCGA glioblastoma WGS dataset in prioritizing survival-associated mutations and regulatory regions. The analysis identified 572 splice-disrupting and 9,837 transcription-factor binding site altering mutations occurring in greater than 10% of glioblastoma samples. Survival analysis linked 1352 mutations and 563 disrupted regulatory regions to patient outcomes, enabling stratification via non-coding mutation signatures. All the code, fine-tuned models, and an interactive data portal are publicly available.
Ancient Script Image Recognition and Processing: A Review
Jun 24, 2025Abstract:Ancient scripts, e.g., Egyptian hieroglyphs, Oracle Bone Inscriptions, and Ancient Greek inscriptions, serve as vital carriers of human civilization, embedding invaluable historical and cultural information. Automating ancient script image recognition has gained importance, enabling large-scale interpretation and advancing research in archaeology and digital humanities. With the rise of deep learning, this field has progressed rapidly, with numerous script-specific datasets and models proposed. While these scripts vary widely, spanning phonographic systems with limited glyphs to logographic systems with thousands of complex symbols, they share common challenges and methodological overlaps. Moreover, ancient scripts face unique challenges, including imbalanced data distribution and image degradation, which have driven the development of various dedicated methods. This survey provides a comprehensive review of ancient script image recognition methods. We begin by categorizing existing studies based on script types and analyzing respective recognition methods, highlighting both their differences and shared strategies. We then focus on challenges unique to ancient scripts, systematically examining their impact and reviewing recent solutions, including few-shot learning and noise-robust techniques. Finally, we summarize current limitations and outline promising future directions. Our goal is to offer a structured, forward-looking perspective to support ongoing advancements in the recognition, interpretation, and decipherment of ancient scripts.
Fast and Low-Cost Genomic Foundation Models via Outlier Removal
May 01, 2025Abstract:We propose the first unified adversarial attack benchmark for Genomic Foundation Models (GFMs), named GERM. Unlike existing GFM benchmarks, GERM offers the first comprehensive evaluation framework to systematically assess the vulnerability of GFMs to adversarial attacks. Methodologically, we evaluate the adversarial robustness of five state-of-the-art GFMs using four widely adopted attack algorithms and three defense strategies. Importantly, our benchmark provides an accessible and comprehensive framework to analyze GFM vulnerabilities with respect to model architecture, quantization schemes, and training datasets. Empirically, transformer-based models exhibit greater robustness to adversarial perturbations compared to HyenaDNA, highlighting the impact of architectural design on vulnerability. Moreover, adversarial attacks frequently target biologically significant genomic regions, suggesting that these models effectively capture meaningful sequence features.
Learning to Instruct for Visual Instruction Tuning
Mar 28, 2025Abstract:We propose LIT, an advancement of visual instruction tuning (VIT). While VIT equips Multimodal LLMs (MLLMs) with promising multimodal capabilities, the current design choices for VIT often result in overfitting and shortcut learning, potentially degrading performance. This gap arises from an overemphasis on instruction-following abilities, while neglecting the proactive understanding of visual information. Inspired by this, LIT adopts a simple yet effective approach by incorporating the loss function into both the instruction and response sequences. It seamlessly expands the training data, and regularizes the MLLMs from overly relying on language priors. Based on this merit, LIT achieves a significant relative improvement of up to 9% on comprehensive multimodal benchmarks, requiring no additional training data and incurring negligible computational overhead. Surprisingly, LIT attains exceptional fundamental visual capabilities, yielding up to an 18% improvement in captioning performance, while simultaneously alleviating hallucination in MLLMs.
USE: Dynamic User Modeling with Stateful Sequence Models
Mar 20, 2024



Abstract:User embeddings play a crucial role in user engagement forecasting and personalized services. Recent advances in sequence modeling have sparked interest in learning user embeddings from behavioral data. Yet behavior-based user embedding learning faces the unique challenge of dynamic user modeling. As users continuously interact with the apps, user embeddings should be periodically updated to account for users' recent and long-term behavior patterns. Existing methods highly rely on stateless sequence models that lack memory of historical behavior. They have to either discard historical data and use only the most recent data or reprocess the old and new data jointly. Both cases incur substantial computational overhead. To address this limitation, we introduce User Stateful Embedding (USE). USE generates user embeddings and reflects users' evolving behaviors without the need for exhaustive reprocessing by storing previous model states and revisiting them in the future. Furthermore, we introduce a novel training objective named future W-behavior prediction to transcend the limitations of next-token prediction by forecasting a broader horizon of upcoming user behaviors. By combining it with the Same User Prediction, a contrastive learning-based objective that predicts whether different segments of behavior sequences belong to the same user, we further improve the embeddings' distinctiveness and representativeness. We conducted experiments on 8 downstream tasks using Snapchat users' behavioral logs in both static (i.e., fixed user behavior sequences) and dynamic (i.e., periodically updated user behavior sequences) settings. We demonstrate USE's superior performance over established baselines. The results underscore USE's effectiveness and efficiency in integrating historical and recent user behavior sequences into user embeddings in dynamic user modeling.
DNABERT-S: Learning Species-Aware DNA Embedding with Genome Foundation Models
Feb 15, 2024



Abstract:Effective DNA embedding remains crucial in genomic analysis, particularly in scenarios lacking labeled data for model fine-tuning, despite the significant advancements in genome foundation models. A prime example is metagenomics binning, a critical process in microbiome research that aims to group DNA sequences by their species from a complex mixture of DNA sequences derived from potentially thousands of distinct, often uncharacterized species. To fill the lack of effective DNA embedding models, we introduce DNABERT-S, a genome foundation model that specializes in creating species-aware DNA embeddings. To encourage effective embeddings to error-prone long-read DNA sequences, we introduce Manifold Instance Mixup (MI-Mix), a contrastive objective that mixes the hidden representations of DNA sequences at randomly selected layers and trains the model to recognize and differentiate these mixed proportions at the output layer. We further enhance it with the proposed Curriculum Contrastive Learning (C$^2$LR) strategy. Empirical results on 18 diverse datasets showed DNABERT-S's remarkable performance. It outperforms the top baseline's performance in 10-shot species classification with just a 2-shot training while doubling the Adjusted Rand Index (ARI) in species clustering and substantially increasing the number of correctly identified species in metagenomics binning. The code, data, and pre-trained model are publicly available at https://github.com/Zhihan1996/DNABERT_S.
Designing and Evaluating General-Purpose User Representations Based on Behavioral Logs from a Measurement Process Perspective: A Case Study with Snapchat
Dec 19, 2023Abstract:In human-computer interaction, understanding user behaviors and tailoring systems accordingly is pivotal. To this end, general-purpose user representation learning based on behavior logs is emerging as a powerful tool in user modeling, offering adaptability to various downstream tasks such as item recommendations and ad conversion prediction, without the need to fine-tune the upstream user model. While this methodology has shown promise in contexts like search engines and e-commerce platforms, its fit for instant messaging apps, a cornerstone of modern digital communication, remains largely uncharted. These apps, with their distinct interaction patterns, data structures, and user expectations, necessitate specialized attention. We explore this user modeling approach with Snapchat data as a case study. Furthermore, we introduce a novel design and evaluation framework rooted in the principles of the Measurement Process Framework from social science research methodology. Using this new framework, we design a Transformer-based user model that can produce high-quality general-purpose user representations for instant messaging platforms like Snapchat.
Combating Representation Learning Disparity with Geometric Harmonization
Oct 26, 2023



Abstract:Self-supervised learning (SSL) as an effective paradigm of representation learning has achieved tremendous success on various curated datasets in diverse scenarios. Nevertheless, when facing the long-tailed distribution in real-world applications, it is still hard for existing methods to capture transferable and robust representation. Conventional SSL methods, pursuing sample-level uniformity, easily leads to representation learning disparity where head classes dominate the feature regime but tail classes passively collapse. To address this problem, we propose a novel Geometric Harmonization (GH) method to encourage category-level uniformity in representation learning, which is more benign to the minority and almost does not hurt the majority under long-tailed distribution. Specially, GH measures the population statistics of the embedding space on top of self-supervised learning, and then infer an fine-grained instance-wise calibration to constrain the space expansion of head classes and avoid the passive collapse of tail classes. Our proposal does not alter the setting of SSL and can be easily integrated into existing methods in a low-cost manner. Extensive results on a range of benchmark datasets show the effectiveness of GH with high tolerance to the distribution skewness. Our code is available at https://github.com/MediaBrain-SJTU/Geometric-Harmonization.
Efficient Action Robust Reinforcement Learning with Probabilistic Policy Execution Uncertainty
Jul 20, 2023
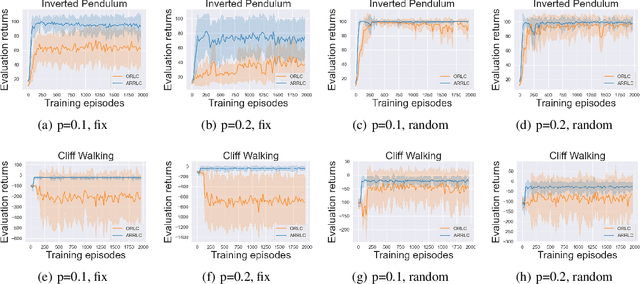
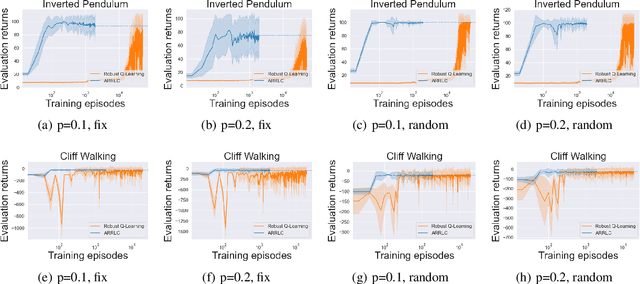
Abstract:Robust reinforcement learning (RL) aims to find a policy that optimizes the worst-case performance in the face of uncertainties. In this paper, we focus on action robust RL with the probabilistic policy execution uncertainty, in which, instead of always carrying out the action specified by the policy, the agent will take the action specified by the policy with probability $1-\rho$ and an alternative adversarial action with probability $\rho$. We establish the existence of an optimal policy on the action robust MDPs with probabilistic policy execution uncertainty and provide the action robust Bellman optimality equation for its solution. Furthermore, we develop Action Robust Reinforcement Learning with Certificates (ARRLC) algorithm that achieves minimax optimal regret and sample complexity. Furthermore, we conduct numerical experiments to validate our approach's robustness, demonstrating that ARRLC outperforms non-robust RL algorithms and converges faster than the robust TD algorithm in the presence of action perturbations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge