Jiayi Wang
Computer Science and Engineering, University of California San Diego, San Diego, USA
MIRROR: A Multi-Agent Framework with Iterative Adaptive Revision and Hierarchical Retrieval for Optimization Modeling in Operations Research
Feb 03, 2026Abstract:Operations Research (OR) relies on expert-driven modeling-a slow and fragile process ill-suited to novel scenarios. While large language models (LLMs) can automatically translate natural language into optimization models, existing approaches either rely on costly post-training or employ multi-agent frameworks, yet most still lack reliable collaborative error correction and task-specific retrieval, often leading to incorrect outputs. We propose MIRROR, a fine-tuning-free, end-to-end multi-agent framework that directly translates natural language optimization problems into mathematical models and solver code. MIRROR integrates two core mechanisms: (1) execution-driven iterative adaptive revision for automatic error correction, and (2) hierarchical retrieval to fetch relevant modeling and coding exemplars from a carefully curated exemplar library. Experiments show that MIRROR outperforms existing methods on standard OR benchmarks, with notable results on complex industrial datasets such as IndustryOR and Mamo-ComplexLP. By combining precise external knowledge infusion with systematic error correction, MIRROR provides non-expert users with an efficient and reliable OR modeling solution, overcoming the fundamental limitations of general-purpose LLMs in expert optimization tasks.
Generation-Augmented Generation: A Plug-and-Play Framework for Private Knowledge Injection in Large Language Models
Jan 13, 2026Abstract:In domains such as biomedicine, materials, and finance, high-stakes deployment of large language models (LLMs) requires injecting private, domain-specific knowledge that is proprietary, fast-evolving, and under-represented in public pretraining. However, the two dominant paradigms for private knowledge injection each have pronounced drawbacks: fine-tuning is expensive to iterate, and continual updates risk catastrophic forgetting and general-capability regression; retrieval-augmented generation (RAG) keeps the base model intact but is brittle in specialized private corpora due to chunk-induced evidence fragmentation, retrieval drift, and long-context pressure that yields query-dependent prompt inflation. Inspired by how multimodal LLMs align heterogeneous modalities into a shared semantic space, we propose Generation-Augmented Generation (GAG), which treats private expertise as an additional expert modality and injects it via a compact, representation-level interface aligned to the frozen base model, avoiding prompt-time evidence serialization while enabling plug-and-play specialization and scalable multi-domain composition with reliable selective activation. Across two private scientific QA benchmarks (immunology adjuvant and catalytic materials) and mixed-domain evaluations, GAG improves specialist performance over strong RAG baselines by 15.34% and 14.86% on the two benchmarks, respectively, while maintaining performance on six open general benchmarks and enabling near-oracle selective activation for scalable multi-domain deployment.
Data Agent: A Holistic Architecture for Orchestrating Data+AI Ecosystems
Jul 02, 2025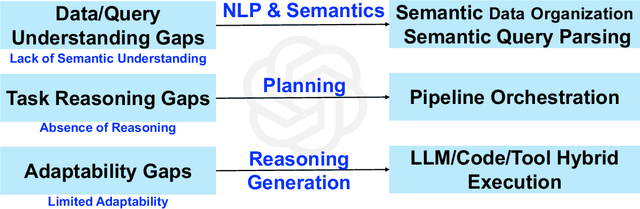
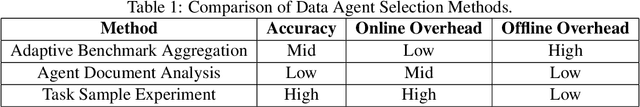
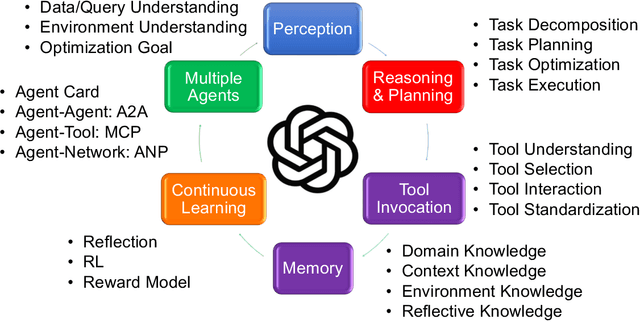
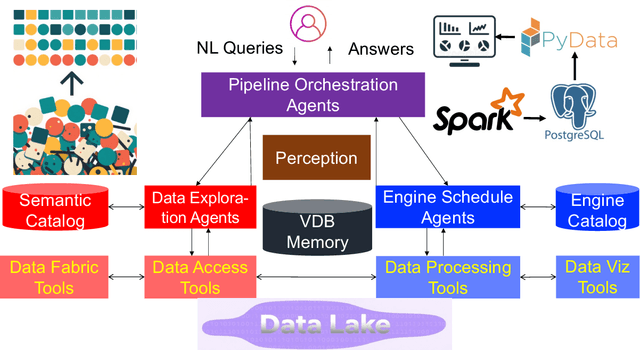
Abstract:Traditional Data+AI systems utilize data-driven techniques to optimize performance, but they rely heavily on human experts to orchestrate system pipelines, enabling them to adapt to changes in data, queries, tasks, and environments. For instance, while there are numerous data science tools available, developing a pipeline planning system to coordinate these tools remains challenging. This difficulty arises because existing Data+AI systems have limited capabilities in semantic understanding, reasoning, and planning. Fortunately, we have witnessed the success of large language models (LLMs) in enhancing semantic understanding, reasoning, and planning abilities. It is crucial to incorporate LLM techniques to revolutionize data systems for orchestrating Data+AI applications effectively. To achieve this, we propose the concept of a 'Data Agent' - a comprehensive architecture designed to orchestrate Data+AI ecosystems, which focuses on tackling data-related tasks by integrating knowledge comprehension, reasoning, and planning capabilities. We delve into the challenges involved in designing data agents, such as understanding data/queries/environments/tools, orchestrating pipelines/workflows, optimizing and executing pipelines, and fostering pipeline self-reflection. Furthermore, we present examples of data agent systems, including a data science agent, data analytics agents (such as unstructured data analytics agent, semantic structured data analytics agent, data lake analytics agent, and multi-modal data analytics agent), and a database administrator (DBA) agent. We also outline several open challenges associated with designing data agent systems.
A Comprehensive Survey on Continual Learning in Generative Models
Jun 16, 2025Abstract:The rapid advancement of generative models has enabled modern AI systems to comprehend and produce highly sophisticated content, even achieving human-level performance in specific domains. However, these models remain fundamentally constrained by catastrophic forgetting - a persistent challenge where adapting to new tasks typically leads to significant degradation in performance on previously learned tasks. To address this practical limitation, numerous approaches have been proposed to enhance the adaptability and scalability of generative models in real-world applications. In this work, we present a comprehensive survey of continual learning methods for mainstream generative models, including large language models, multimodal large language models, vision language action models, and diffusion models. Drawing inspiration from the memory mechanisms of the human brain, we systematically categorize these approaches into three paradigms: architecture-based, regularization-based, and replay-based methods, while elucidating their underlying methodologies and motivations. We further analyze continual learning setups for different generative models, including training objectives, benchmarks, and core backbones, offering deeper insights into the field. The project page of this paper is available at https://github.com/Ghy0501/Awesome-Continual-Learning-in-Generative-Models.
SSA-COMET: Do LLMs Outperform Learned Metrics in Evaluating MT for Under-Resourced African Languages?
Jun 05, 2025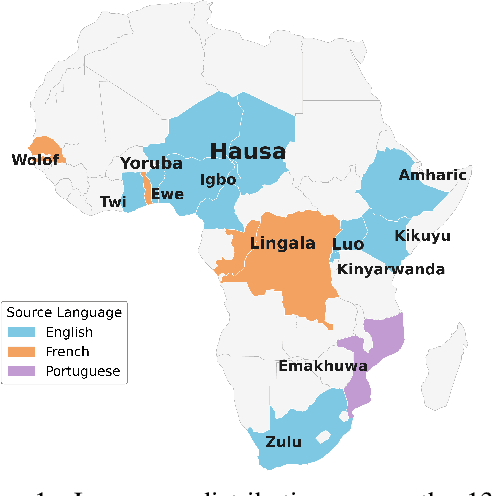
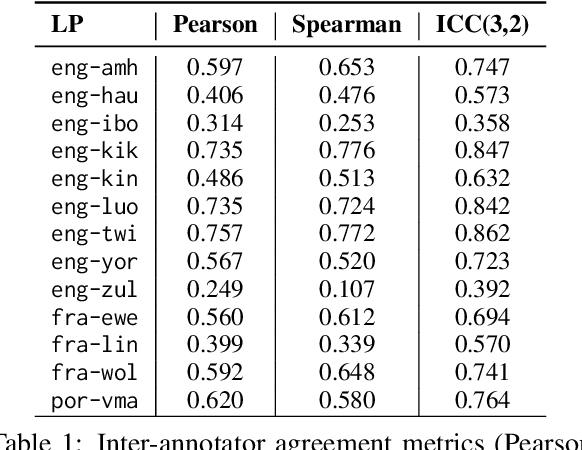
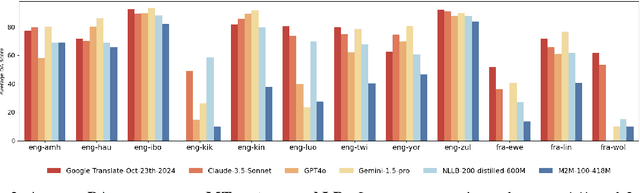
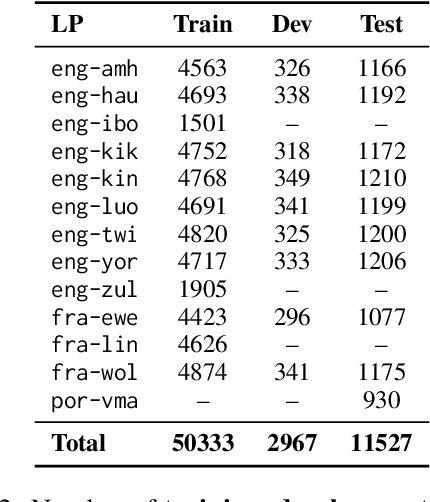
Abstract:Evaluating machine translation (MT) quality for under-resourced African languages remains a significant challenge, as existing metrics often suffer from limited language coverage and poor performance in low-resource settings. While recent efforts, such as AfriCOMET, have addressed some of the issues, they are still constrained by small evaluation sets, a lack of publicly available training data tailored to African languages, and inconsistent performance in extremely low-resource scenarios. In this work, we introduce SSA-MTE, a large-scale human-annotated MT evaluation (MTE) dataset covering 13 African language pairs from the News domain, with over 63,000 sentence-level annotations from a diverse set of MT systems. Based on this data, we develop SSA-COMET and SSA-COMET-QE, improved reference-based and reference-free evaluation metrics. We also benchmark prompting-based approaches using state-of-the-art LLMs like GPT-4o and Claude. Our experimental results show that SSA-COMET models significantly outperform AfriCOMET and are competitive with the strongest LLM (Gemini 2.5 Pro) evaluated in our study, particularly on low-resource languages such as Twi, Luo, and Yoruba. All resources are released under open licenses to support future research.
RSFAKE-1M: A Large-Scale Dataset for Detecting Diffusion-Generated Remote Sensing Forgeries
May 29, 2025



Abstract:Detecting forged remote sensing images is becoming increasingly critical, as such imagery plays a vital role in environmental monitoring, urban planning, and national security. While diffusion models have emerged as the dominant paradigm for image generation, their impact on remote sensing forgery detection remains underexplored. Existing benchmarks primarily target GAN-based forgeries or focus on natural images, limiting progress in this critical domain. To address this gap, we introduce RSFAKE-1M, a large-scale dataset of 500K forged and 500K real remote sensing images. The fake images are generated by ten diffusion models fine-tuned on remote sensing data, covering six generation conditions such as text prompts, structural guidance, and inpainting. This paper presents the construction of RSFAKE-1M along with a comprehensive experimental evaluation using both existing detectors and unified baselines. The results reveal that diffusion-based remote sensing forgeries remain challenging for current methods, and that models trained on RSFAKE-1M exhibit notably improved generalization and robustness. Our findings underscore the importance of RSFAKE-1M as a foundation for developing and evaluating next-generation forgery detection approaches in the remote sensing domain. The dataset and other supplementary materials are available at https://huggingface.co/datasets/TZHSW/RSFAKE/.
ReasoningShield: Content Safety Detection over Reasoning Traces of Large Reasoning Models
May 22, 2025Abstract:Large Reasoning Models (LRMs) are transforming the AI landscape with advanced reasoning capabilities. While the generated reasoning traces enhance model transparency, they can still contain unsafe content, even when the final answer appears safe. Existing moderation tools, primarily designed for question-answer (QA) pairs, are empirically ineffective at detecting hidden risks embedded in reasoning traces. After identifying the key challenges, we formally define the question-thought (QT) moderation task and propose ReasoningShield, the first safety detection model tailored to identify potential risks in the reasoning trace before reaching the final answer. To construct the model, we synthesize a high-quality reasoning safety detection dataset comprising over 8,000 question-thought pairs spanning ten risk categories and three safety levels. Our dataset construction process incorporates a comprehensive human-AI collaborative annotation pipeline, which achieves over 93% annotation accuracy while significantly reducing human costs. On a diverse set of in-distribution and out-of-distribution benchmarks, ReasoningShield outperforms mainstream content safety moderation models in identifying risks within reasoning traces, with an average F1 score exceeding 0.92. Notably, despite being trained on our QT dataset only, ReasoningShield also demonstrates competitive performance in detecting unsafe question-answer pairs on traditional benchmarks, rivaling baselines trained on 10 times larger datasets and base models, which strongly validates the quality of our dataset. Furthermore, ReasoningShield is built upon compact 1B/3B base models to facilitate lightweight deployment and provides human-friendly risk analysis by default. To foster future research, we publicly release all the resources.
Prototype-Based Information Compensation Network for Multi-Source Remote Sensing Data Classification
May 06, 2025Abstract:Multi-source remote sensing data joint classification aims to provide accuracy and reliability of land cover classification by leveraging the complementary information from multiple data sources. Existing methods confront two challenges: inter-frequency multi-source feature coupling and inconsistency of complementary information exploration. To solve these issues, we present a Prototype-based Information Compensation Network (PICNet) for land cover classification based on HSI and SAR/LiDAR data. Specifically, we first design a frequency interaction module to enhance the inter-frequency coupling in multi-source feature extraction. The multi-source features are first decoupled into high- and low-frequency components. Then, these features are recoupled to achieve efficient inter-frequency communication. Afterward, we design a prototype-based information compensation module to model the global multi-source complementary information. Two sets of learnable modality prototypes are introduced to represent the global modality information of multi-source data. Subsequently, cross-modal feature integration and alignment are achieved through cross-attention computation between the modality-specific prototype vectors and the raw feature representations. Extensive experiments on three public datasets demonstrate the significant superiority of our PICNet over state-of-the-art methods. The codes are available at https://github.com/oucailab/PICNet.
A comparative analysis of rank aggregation methods for the partial label ranking problem
Feb 25, 2025



Abstract:The label ranking problem is a supervised learning scenario in which the learner predicts a total order of the class labels for a given input instance. Recently, research has increasingly focused on the partial label ranking problem, a generalization of the label ranking problem that allows ties in the predicted orders. So far, most existing learning approaches for the partial label ranking problem rely on approximation algorithms for rank aggregation in the final prediction step. This paper explores several alternative aggregation methods for this critical step, including scoring-based and probabilistic-based rank aggregation approaches. To enhance their suitability for the more general partial label ranking problem, the investigated methods are extended to increase the likelihood of producing ties. Experimental evaluations on standard benchmarks demonstrate that scoring-based variants consistently outperform the current state-of-the-art method in handling incomplete information. In contrast, probabilistic-based variants fail to achieve competitive performance.
Multilingual Language Model Pretraining using Machine-translated Data
Feb 18, 2025Abstract:High-resource languages such as English, enables the pretraining of high-quality large language models (LLMs). The same can not be said for most other languages as LLMs still underperform for non-English languages, likely due to a gap in the quality and diversity of the available multilingual pretraining corpora. In this work, we find that machine-translated texts from a single high-quality source language can contribute significantly to the pretraining quality of multilingual LLMs. We translate FineWeb-Edu, a high-quality English web dataset, into nine languages, resulting in a 1.7-trillion-token dataset, which we call TransWebEdu and pretrain a 1.3B-parameter model, TransWebLLM, from scratch on this dataset. Across nine non-English reasoning tasks, we show that TransWebLLM matches or outperforms state-of-the-art multilingual models trained using closed data, such as Llama3.2, Qwen2.5, and Gemma, despite using an order of magnitude less data. We demonstrate that adding less than 5% of TransWebEdu as domain-specific pretraining data sets a new state-of-the-art in Arabic, Italian, Indonesian, Swahili, and Welsh understanding and commonsense reasoning tasks. To promote reproducibility, we release our corpus, models, and training pipeline under Open Source Initiative-approved licenses.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge