Meng Zhou
A Multi-Dimensional Constraint Framework for Evaluating and Improving Instruction Following in Large Language Models
May 12, 2025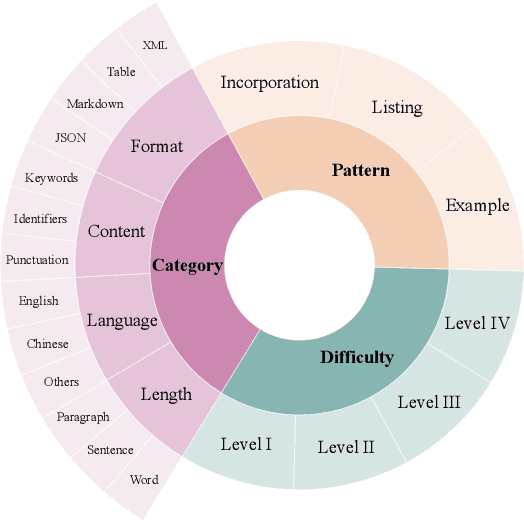
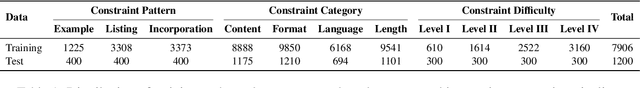
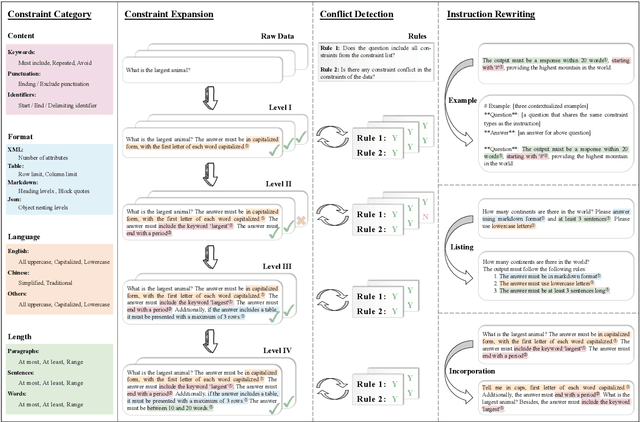
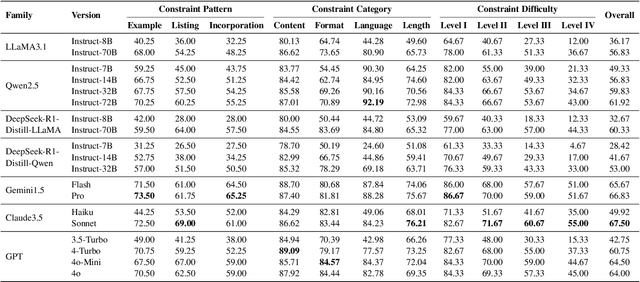
Abstract:Instruction following evaluates large language models (LLMs) on their ability to generate outputs that adhere to user-defined constraints. However, existing benchmarks often rely on templated constraint prompts, which lack the diversity of real-world usage and limit fine-grained performance assessment. To fill this gap, we propose a multi-dimensional constraint framework encompassing three constraint patterns, four constraint categories, and four difficulty levels. Building on this framework, we develop an automated instruction generation pipeline that performs constraint expansion, conflict detection, and instruction rewriting, yielding 1,200 code-verifiable instruction-following test samples. We evaluate 19 LLMs across seven model families and uncover substantial variation in performance across constraint forms. For instance, average performance drops from 77.67% at Level I to 32.96% at Level IV. Furthermore, we demonstrate the utility of our approach by using it to generate data for reinforcement learning, achieving substantial gains in instruction following without degrading general performance. In-depth analysis indicates that these gains stem primarily from modifications in the model's attention modules parameters, which enhance constraint recognition and adherence. Code and data are available in https://github.com/Junjie-Ye/MulDimIF.
Dynamic Attention Mechanism in Spatiotemporal Memory Networks for Object Tracking
Mar 21, 2025



Abstract:Mainstream visual object tracking frameworks predominantly rely on template matching paradigms. Their performance heavily depends on the quality of template features, which becomes increasingly challenging to maintain in complex scenarios involving target deformation, occlusion, and background clutter. While existing spatiotemporal memory-based trackers emphasize memory capacity expansion, they lack effective mechanisms for dynamic feature selection and adaptive fusion. To address this gap, we propose a Dynamic Attention Mechanism in Spatiotemporal Memory Network (DASTM) with two key innovations: 1) A differentiable dynamic attention mechanism that adaptively adjusts channel-spatial attention weights by analyzing spatiotemporal correlations between the templates and memory features; 2) A lightweight gating network that autonomously allocates computational resources based on target motion states, prioritizing high-discriminability features in challenging scenarios. Extensive evaluations on OTB-2015, VOT 2018, LaSOT, and GOT-10K benchmarks demonstrate our DASTM's superiority, achieving state-of-the-art performance in success rate, robustness, and real-time efficiency, thereby offering a novel solution for real-time tracking in complex environments.
Efficient Parallel Genetic Algorithm for Perturbed Substructure Optimization in Complex Network
Dec 30, 2024



Abstract:Evolutionary computing, particularly genetic algorithm (GA), is a combinatorial optimization method inspired by natural selection and the transmission of genetic information, which is widely used to identify optimal solutions to complex problems through simulated programming and iteration. Due to its strong adaptability, flexibility, and robustness, GA has shown significant performance and potentiality on perturbed substructure optimization (PSSO), an important graph mining problem that achieves its goals by modifying network structures. However, the efficiency and practicality of GA-based PSSO face enormous challenges due to the complexity and diversity of application scenarios. While some research has explored acceleration frameworks in evolutionary computing, their performance on PSSO remains limited due to a lack of scenario generalizability. Based on these, this paper is the first to present the GA-based PSSO Acceleration framework (GAPA), which simplifies the GA development process and supports distributed acceleration. Specifically, it reconstructs the genetic operation and designs a development framework for efficient parallel acceleration. Meanwhile, GAPA includes an extensible library that optimizes and accelerates 10 PSSO algorithms, covering 4 crucial tasks for graph mining. Comprehensive experiments on 18 datasets across 4 tasks and 10 algorithms effectively demonstrate the superiority of GAPA, achieving an average of 4x the acceleration of Evox. The repository is in https://github.com/NetAlsGroup/GAPA.
Training Agents with Weakly Supervised Feedback from Large Language Models
Nov 29, 2024



Abstract:Large Language Models (LLMs) offer a promising basis for creating agents that can tackle complex tasks through iterative environmental interaction. Existing methods either require these agents to mimic expert-provided trajectories or rely on definitive environmental feedback for reinforcement learning which limits their application to specific scenarios like gaming or code generation. This paper introduces a novel training method for LLM-based agents using weakly supervised signals from a critic LLM, bypassing the need for expert trajectories or definitive feedback. Our agents are trained in iterative manner, where they initially generate trajectories through environmental interaction. Subsequently, a critic LLM selects a subset of good trajectories, which are then used to update the agents, enabling them to generate improved trajectories in the next iteration. Extensive tests on the API-bank dataset show consistent improvement in our agents' capabilities and comparable performance to GPT-4, despite using open-source models with much fewer parameters.
Edge-Enhanced Dilated Residual Attention Network for Multimodal Medical Image Fusion
Nov 18, 2024



Abstract:Multimodal medical image fusion is a crucial task that combines complementary information from different imaging modalities into a unified representation, thereby enhancing diagnostic accuracy and treatment planning. While deep learning methods, particularly Convolutional Neural Networks (CNNs) and Transformers, have significantly advanced fusion performance, some of the existing CNN-based methods fall short in capturing fine-grained multiscale and edge features, leading to suboptimal feature integration. Transformer-based models, on the other hand, are computationally intensive in both the training and fusion stages, making them impractical for real-time clinical use. Moreover, the clinical application of fused images remains unexplored. In this paper, we propose a novel CNN-based architecture that addresses these limitations by introducing a Dilated Residual Attention Network Module for effective multiscale feature extraction, coupled with a gradient operator to enhance edge detail learning. To ensure fast and efficient fusion, we present a parameter-free fusion strategy based on the weighted nuclear norm of softmax, which requires no additional computations during training or inference. Extensive experiments, including a downstream brain tumor classification task, demonstrate that our approach outperforms various baseline methods in terms of visual quality, texture preservation, and fusion speed, making it a possible practical solution for real-world clinical applications. The code will be released at https://github.com/simonZhou86/en_dran.
Towards Democratizing Multilingual Large Language Models For Medicine Through A Two-Stage Instruction Fine-tuning Approach
Sep 09, 2024



Abstract:Open-source, multilingual medical large language models (LLMs) have the potential to serve linguistically diverse populations across different regions. Adapting generic LLMs for healthcare often requires continual pretraining, but this approach is computationally expensive and sometimes impractical. Instruction fine-tuning on a specific task may not always guarantee optimal performance due to the lack of broader domain knowledge that the model needs to understand and reason effectively in diverse scenarios. To address these challenges, we introduce two multilingual instruction fine-tuning datasets, MMed-IFT and MMed-IFT-MC, containing over 200k high-quality medical samples in six languages. We propose a two-stage training paradigm: the first stage injects general medical knowledge using MMed-IFT, while the second stage fine-tunes task-specific multiple-choice questions with MMed-IFT-MC. Our method achieves competitive results on both English and multilingual benchmarks, striking a balance between computational efficiency and performance. We plan to make our dataset and model weights public at \url{https://github.com/SpassMed/Med-Llama3} in the future.
A Labeled Ophthalmic Ultrasound Dataset with Medical Report Generation Based on Cross-modal Deep Learning
Jul 26, 2024



Abstract:Ultrasound imaging reveals eye morphology and aids in diagnosing and treating eye diseases. However, interpreting diagnostic reports requires specialized physicians. We present a labeled ophthalmic dataset for the precise analysis and the automated exploration of medical images along with their associated reports. It collects three modal data, including the ultrasound images, blood flow information and examination reports from 2,417 patients at an ophthalmology hospital in Shenyang, China, during the year 2018, in which the patient information is de-identified for privacy protection. To the best of our knowledge, it is the only ophthalmic dataset that contains the three modal information simultaneously. It incrementally consists of 4,858 images with the corresponding free-text reports, which describe 15 typical imaging findings of intraocular diseases and the corresponding anatomical locations. Each image shows three kinds of blood flow indices at three specific arteries, i.e., nine parameter values to describe the spectral characteristics of blood flow distribution. The reports were written by ophthalmologists during the clinical care. The proposed dataset is applied to generate medical report based on the cross-modal deep learning model. The experimental results demonstrate that our dataset is suitable for training supervised models concerning cross-modal medical data.
Generating 3D Brain Tumor Regions in MRI using Vector-Quantization Generative Adversarial Networks
Oct 02, 2023



Abstract:Medical image analysis has significantly benefited from advancements in deep learning, particularly in the application of Generative Adversarial Networks (GANs) for generating realistic and diverse images that can augment training datasets. However, the effectiveness of such approaches is often limited by the amount of available data in clinical settings. Additionally, the common GAN-based approach is to generate entire image volumes, rather than solely the region of interest (ROI). Research on deep learning-based brain tumor classification using MRI has shown that it is easier to classify the tumor ROIs compared to the entire image volumes. In this work, we present a novel framework that uses vector-quantization GAN and a transformer incorporating masked token modeling to generate high-resolution and diverse 3D brain tumor ROIs that can be directly used as augmented data for the classification of brain tumor ROI. We apply our method to two imbalanced datasets where we augment the minority class: (1) the Multimodal Brain Tumor Segmentation Challenge (BraTS) 2019 dataset to generate new low-grade glioma (LGG) ROIs to balance with high-grade glioma (HGG) class; (2) the internal pediatric LGG (pLGG) dataset tumor ROIs with BRAF V600E Mutation genetic marker to balance with BRAF Fusion genetic marker class. We show that the proposed method outperforms various baseline models in both qualitative and quantitative measurements. The generated data was used to balance the data in the brain tumor types classification task. Using the augmented data, our approach surpasses baseline models by 6.4% in AUC on the BraTS 2019 dataset and 4.3% in AUC on our internal pLGG dataset. The results indicate the generated tumor ROIs can effectively address the imbalanced data problem. Our proposed method has the potential to facilitate an accurate diagnosis of rare brain tumors using MRI scans.
Domain Transfer Through Image-to-Image Translation for Uncertainty-Aware Prostate Cancer Classification
Jul 02, 2023Abstract:Prostate Cancer (PCa) is often diagnosed using High-resolution 3.0 Tesla(T) MRI, which has been widely established in clinics. However, there are still many medical centers that use 1.5T MRI units in the actual diagnostic process of PCa. In the past few years, deep learning-based models have been proven to be efficient on the PCa classification task and can be successfully used to support radiologists during the diagnostic process. However, training such models often requires a vast amount of data, and sometimes it is unobtainable in practice. Additionally, multi-source MRIs can pose challenges due to cross-domain distribution differences. In this paper, we have presented a novel approach for unpaired image-to-image translation of prostate mp-MRI for classifying clinically significant PCa, to be applied in data-constrained settings. First, we introduce domain transfer, a novel pipeline to translate unpaired 3.0T multi-parametric prostate MRIs to 1.5T, to increase the number of training data. Second, we estimate the uncertainty of our models through an evidential deep learning approach; and leverage the dataset filtering technique during the training process. Furthermore, we introduce a simple, yet efficient Evidential Focal Loss that incorporates the focal loss with evidential uncertainty to train our model. Our experiments demonstrate that the proposed method significantly improves the Area Under ROC Curve (AUC) by over 20% compared to the previous work (98.4% vs. 76.2%). We envision that providing prediction uncertainty to radiologists may help them focus more on uncertain cases and thus expedite the diagnostic process effectively. Our code is available at https://github.com/med-i-lab/DT_UE_PCa
From Clozing to Comprehending: Retrofitting Pre-trained Language Model to Pre-trained Machine Reader
Dec 09, 2022Abstract:We present Pre-trained Machine Reader (PMR), a novel method to retrofit Pre-trained Language Models (PLMs) into Machine Reading Comprehension (MRC) models without acquiring labeled data. PMR is capable of resolving the discrepancy between model pre-training and downstream fine-tuning of existing PLMs, and provides a unified solver for tackling various extraction tasks. To achieve this, we construct a large volume of general-purpose and high-quality MRC-style training data with the help of Wikipedia hyperlinks and design a Wiki Anchor Extraction task to guide the MRC-style pre-training process. Although conceptually simple, PMR is particularly effective in solving extraction tasks including Extractive Question Answering and Named Entity Recognition, where it shows tremendous improvements over previous approaches especially under low-resource settings. Moreover, viewing sequence classification task as a special case of extraction task in our MRC formulation, PMR is even capable to extract high-quality rationales to explain the classification process, providing more explainability of the predictions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge