Anubhav Bhatti
Beyond LoRA: Exploring Efficient Fine-Tuning Techniques for Time Series Foundational Models
Sep 17, 2024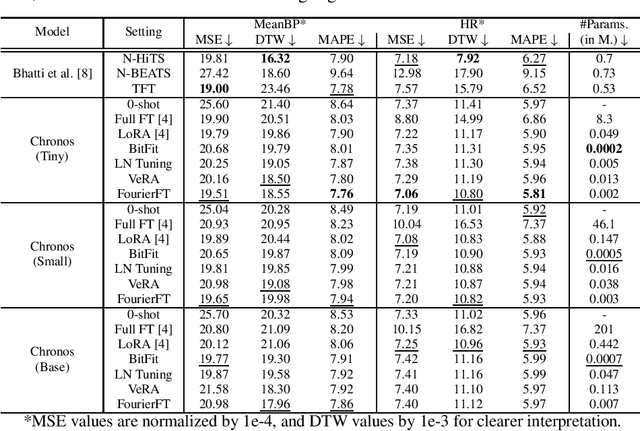
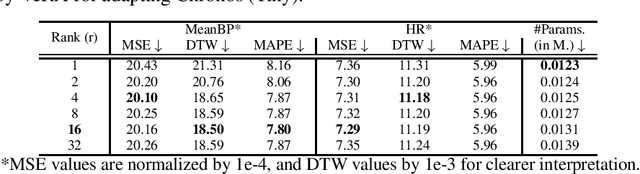
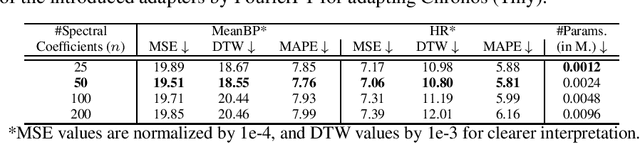

Abstract:Time Series Foundation Models (TSFMs) have recently garnered attention for their ability to model complex, large-scale time series data across domains such as retail, finance, and transportation. However, their application to sensitive, domain-specific fields like healthcare remains challenging, primarily due to the difficulty of fine-tuning these models for specialized, out-of-domain tasks with scarce publicly available datasets. In this work, we explore the use of Parameter-Efficient Fine-Tuning (PEFT) techniques to address these limitations, focusing on healthcare applications, particularly ICU vitals forecasting for sepsis patients. We introduce and evaluate two selective (BitFit and LayerNorm Tuning) and two additive (VeRA and FourierFT) PEFT techniques on multiple configurations of the Chronos TSFM for forecasting vital signs of sepsis patients. Our comparative analysis demonstrates that some of these PEFT methods outperform LoRA in terms of parameter efficiency and domain adaptation, establishing state-of-the-art (SOTA) results in ICU vital forecasting tasks. Interestingly, FourierFT applied to the Chronos (Tiny) variant surpasses the SOTA model while fine-tuning only 2,400 parameters compared to the 700K parameters of the benchmark.
Towards Democratizing Multilingual Large Language Models For Medicine Through A Two-Stage Instruction Fine-tuning Approach
Sep 09, 2024



Abstract:Open-source, multilingual medical large language models (LLMs) have the potential to serve linguistically diverse populations across different regions. Adapting generic LLMs for healthcare often requires continual pretraining, but this approach is computationally expensive and sometimes impractical. Instruction fine-tuning on a specific task may not always guarantee optimal performance due to the lack of broader domain knowledge that the model needs to understand and reason effectively in diverse scenarios. To address these challenges, we introduce two multilingual instruction fine-tuning datasets, MMed-IFT and MMed-IFT-MC, containing over 200k high-quality medical samples in six languages. We propose a two-stage training paradigm: the first stage injects general medical knowledge using MMed-IFT, while the second stage fine-tunes task-specific multiple-choice questions with MMed-IFT-MC. Our method achieves competitive results on both English and multilingual benchmarks, striking a balance between computational efficiency and performance. We plan to make our dataset and model weights public at \url{https://github.com/SpassMed/Med-Llama3} in the future.
Low-Rank Adaptation of Time Series Foundational Models for Out-of-Domain Modality Forecasting
May 16, 2024Abstract:Low-Rank Adaptation (LoRA) is a widely used technique for fine-tuning large pre-trained or foundational models across different modalities and tasks. However, its application to time series data, particularly within foundational models, remains underexplored. This paper examines the impact of LoRA on contemporary time series foundational models: Lag-Llama, MOIRAI, and Chronos. We demonstrate LoRA's fine-tuning potential for forecasting the vital signs of sepsis patients in intensive care units (ICUs), emphasizing the models' adaptability to previously unseen, out-of-domain modalities. Integrating LoRA aims to enhance forecasting performance while reducing inefficiencies associated with fine-tuning large models on limited domain-specific data. Our experiments show that LoRA fine-tuning of time series foundational models significantly improves forecasting, achieving results comparable to state-of-the-art models trained from scratch on similar modalities. We conduct comprehensive ablation studies to demonstrate the trade-offs between the number of tunable parameters and forecasting performance and assess the impact of varying LoRA matrix ranks on model performance.
Interpretable Vital Sign Forecasting with Model Agnostic Attention Maps
May 06, 2024Abstract:Sepsis is a leading cause of mortality in intensive care units (ICUs), representing a substantial medical challenge. The complexity of analyzing diverse vital signs to predict sepsis further aggravates this issue. While deep learning techniques have been advanced for early sepsis prediction, their 'black-box' nature obscures the internal logic, impairing interpretability in critical settings like ICUs. This paper introduces a framework that combines a deep learning model with an attention mechanism that highlights the critical time steps in the forecasting process, thus improving model interpretability and supporting clinical decision-making. We show that the attention mechanism could be adapted to various black box time series forecasting models such as N-HiTS and N-BEATS. Our method preserves the accuracy of conventional deep learning models while enhancing interpretability through attention-weight-generated heatmaps. We evaluated our model on the eICU-CRD dataset, focusing on forecasting vital signs for sepsis patients. We assessed its performance using mean squared error (MSE) and dynamic time warping (DTW) metrics. We explored the attention maps of N-HiTS and N-BEATS, examining the differences in their performance and identifying crucial factors influencing vital sign forecasting.
CLARE: Cognitive Load Assessment in REaltime with Multimodal Data
Apr 26, 2024



Abstract:We present a novel multimodal dataset for Cognitive Load Assessment in REaltime (CLARE). The dataset contains physiological and gaze data from 24 participants with self-reported cognitive load scores as ground-truth labels. The dataset consists of four modalities, namely, Electrocardiography (ECG), Electrodermal Activity (EDA), Electroencephalogram (EEG), and Gaze tracking. To map diverse levels of mental load on participants during experiments, each participant completed four nine-minutes sessions on a computer-based operator performance and mental workload task (the MATB-II software) with varying levels of complexity in one minute segments. During the experiment, participants reported their cognitive load every 10 seconds. For the dataset, we also provide benchmark binary classification results with machine learning and deep learning models on two different evaluation schemes, namely, 10-fold and leave-one-subject-out (LOSO) cross-validation. Benchmark results show that for 10-fold evaluation, the convolutional neural network (CNN) based deep learning model achieves the best classification performance with ECG, EDA, and Gaze. In contrast, for LOSO, the best performance is achieved by the deep learning model with ECG, EDA, and EEG.
SM70: A Large Language Model for Medical Devices
Dec 12, 2023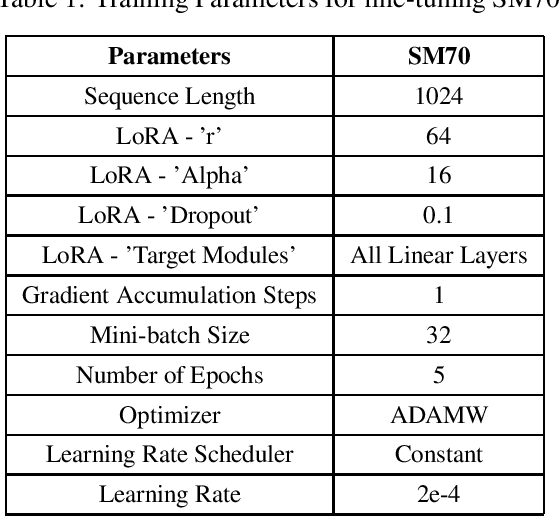
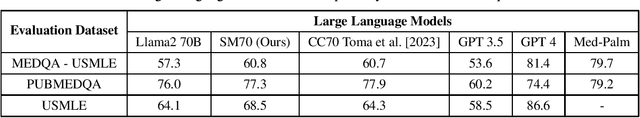
Abstract:We are introducing SM70, a 70 billion-parameter Large Language Model that is specifically designed for SpassMed's medical devices under the brand name 'JEE1' (pronounced as G1 and means 'Life'). This large language model provides more accurate and safe responses to medical-domain questions. To fine-tune SM70, we used around 800K data entries from the publicly available dataset MedAlpaca. The Llama2 70B open-sourced model served as the foundation for SM70, and we employed the QLoRA technique for fine-tuning. The evaluation is conducted across three benchmark datasets - MEDQA - USMLE, PUBMEDQA, and USMLE - each representing a unique aspect of medical knowledge and reasoning. The performance of SM70 is contrasted with other notable LLMs, including Llama2 70B, Clinical Camel 70 (CC70), GPT 3.5, GPT 4, and Med-Palm, to provide a comparative understanding of its capabilities within the medical domain. Our results indicate that SM70 outperforms several established models in these datasets, showcasing its proficiency in handling a range of medical queries, from fact-based questions derived from PubMed abstracts to complex clinical decision-making scenarios. The robust performance of SM70, particularly in the USMLE and PUBMEDQA datasets, suggests its potential as an effective tool in clinical decision support and medical information retrieval. Despite its promising results, the paper also acknowledges the areas where SM70 lags behind the most advanced model, GPT 4, thereby highlighting the need for further development, especially in tasks demanding extensive medical knowledge and intricate reasoning.
Vital Sign Forecasting for Sepsis Patients in ICUs
Nov 08, 2023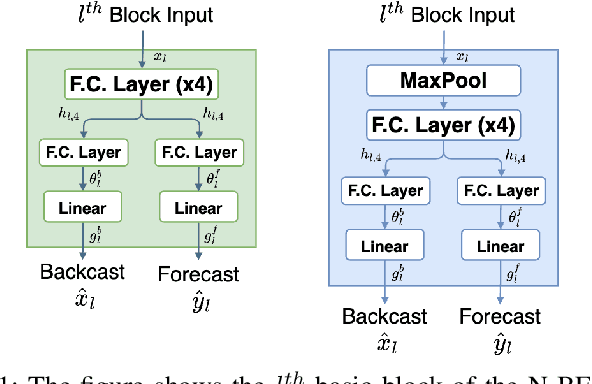
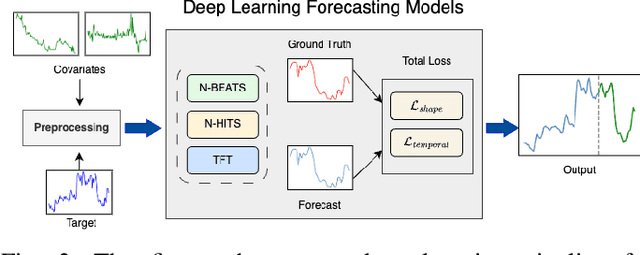
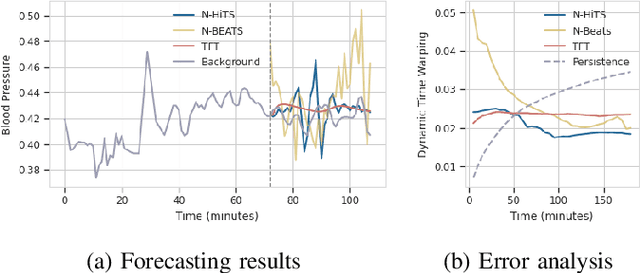
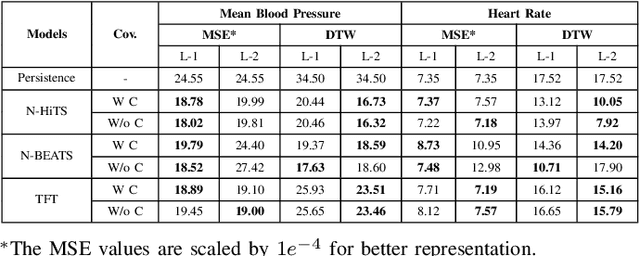
Abstract:Sepsis and septic shock are a critical medical condition affecting millions globally, with a substantial mortality rate. This paper uses state-of-the-art deep learning (DL) architectures to introduce a multi-step forecasting system to predict vital signs indicative of septic shock progression in Intensive Care Units (ICUs). Our approach utilizes a short window of historical vital sign data to forecast future physiological conditions. We introduce a DL-based vital sign forecasting system that predicts up to 3 hours of future vital signs from 6 hours of past data. We further adopt the DILATE loss function to capture better the shape and temporal dynamics of vital signs, which are critical for clinical decision-making. We compare three DL models, N-BEATS, N-HiTS, and Temporal Fusion Transformer (TFT), using the publicly available eICU Collaborative Research Database (eICU-CRD), highlighting their forecasting capabilities in a critical care setting. We evaluate the performance of our models using mean squared error (MSE) and dynamic time warping (DTW) metrics. Our findings show that while TFT excels in capturing overall trends, N-HiTS is superior in retaining short-term fluctuations within a predefined range. This paper demonstrates the potential of deep learning in transforming the monitoring systems in ICUs, potentially leading to significant improvements in patient care and outcomes by accurately forecasting vital signs to assist healthcare providers in detecting early signs of physiological instability and anticipating septic shock.
Interpreting Forecasted Vital Signs Using N-BEATS in Sepsis Patients
Jun 24, 2023

Abstract:Detecting and predicting septic shock early is crucial for the best possible outcome for patients. Accurately forecasting the vital signs of patients with sepsis provides valuable insights to clinicians for timely interventions, such as administering stabilizing drugs or optimizing infusion strategies. Our research examines N-BEATS, an interpretable deep-learning forecasting model that can forecast 3 hours of vital signs for sepsis patients in intensive care units (ICUs). In this work, we use the N-BEATS interpretable configuration to forecast the vital sign trends and compare them with the actual trend to understand better the patient's changing condition and the effects of infused drugs on their vital signs. We evaluate our approach using the publicly available eICU Collaborative Research Database dataset and rigorously evaluate the vital sign forecasts using out-of-sample evaluation criteria. We present the performance of our model using error metrics, including mean squared error (MSE), mean average percentage error (MAPE), and dynamic time warping (DTW), where the best scores achieved are 18.52e-4, 7.60, and 17.63e-3, respectively. We analyze the samples where the forecasted trend does not match the actual trend and study the impact of infused drugs on changing the actual vital signs compared to the forecasted trend. Additionally, we examined the mortality rates of patients where the actual trend and the forecasted trend did not match. We observed that the mortality rate was higher (92%) when the actual and forecasted trends closely matched, compared to when they were not similar (84%).
Multimodal Brain-Computer Interface for In-Vehicle Driver Cognitive Load Measurement: Dataset and Baselines
Apr 09, 2023

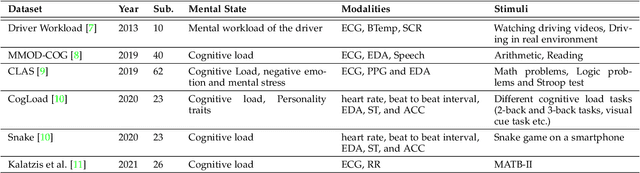
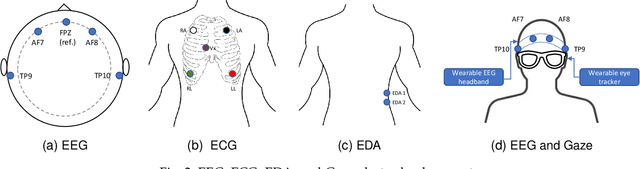
Abstract:Through this paper, we introduce a novel driver cognitive load assessment dataset, CL-Drive, which contains Electroencephalogram (EEG) signals along with other physiological signals such as Electrocardiography (ECG) and Electrodermal Activity (EDA) as well as eye tracking data. The data was collected from 21 subjects while driving in an immersive vehicle simulator, in various driving conditions, to induce different levels of cognitive load in the subjects. The tasks consisted of 9 complexity levels for 3 minutes each. Each driver reported their subjective cognitive load every 10 seconds throughout the experiment. The dataset contains the subjective cognitive load recorded as ground truth. In this paper, we also provide benchmark classification results for different machine learning and deep learning models for both binary and ternary label distributions. We followed 2 evaluation criteria namely 10-fold and leave-one-subject-out (LOSO). We have trained our models on both hand-crafted features as well as on raw data.
AttX: Attentive Cross-Connections for Fusion of Wearable Signals in Emotion Recognition
Jun 09, 2022



Abstract:We propose cross-modal attentive connections, a new dynamic and effective technique for multimodal representation learning from wearable data. Our solution can be integrated into any stage of the pipeline, i.e., after any convolutional layer or block, to create intermediate connections between individual streams responsible for processing each modality. Additionally, our method benefits from two properties. First, it can share information uni-directionally (from one modality to the other) or bi-directionally. Second, it can be integrated into multiple stages at the same time to further allow network gradients to be exchanged in several touch-points. We perform extensive experiments on three public multimodal wearable datasets, WESAD, SWELL-KW, and CASE, and demonstrate that our method can effectively regulate and share information between different modalities to learn better representations. Our experiments further demonstrate that once integrated into simple CNN-based multimodal solutions (2, 3, or 4 modalities), our method can result in superior or competitive performance to state-of-the-art and outperform a variety of baseline uni-modal and classical multimodal methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge