San Lee
Low-Rank Adaptation of Time Series Foundational Models for Out-of-Domain Modality Forecasting
May 16, 2024Abstract:Low-Rank Adaptation (LoRA) is a widely used technique for fine-tuning large pre-trained or foundational models across different modalities and tasks. However, its application to time series data, particularly within foundational models, remains underexplored. This paper examines the impact of LoRA on contemporary time series foundational models: Lag-Llama, MOIRAI, and Chronos. We demonstrate LoRA's fine-tuning potential for forecasting the vital signs of sepsis patients in intensive care units (ICUs), emphasizing the models' adaptability to previously unseen, out-of-domain modalities. Integrating LoRA aims to enhance forecasting performance while reducing inefficiencies associated with fine-tuning large models on limited domain-specific data. Our experiments show that LoRA fine-tuning of time series foundational models significantly improves forecasting, achieving results comparable to state-of-the-art models trained from scratch on similar modalities. We conduct comprehensive ablation studies to demonstrate the trade-offs between the number of tunable parameters and forecasting performance and assess the impact of varying LoRA matrix ranks on model performance.
Interpretable Vital Sign Forecasting with Model Agnostic Attention Maps
May 06, 2024Abstract:Sepsis is a leading cause of mortality in intensive care units (ICUs), representing a substantial medical challenge. The complexity of analyzing diverse vital signs to predict sepsis further aggravates this issue. While deep learning techniques have been advanced for early sepsis prediction, their 'black-box' nature obscures the internal logic, impairing interpretability in critical settings like ICUs. This paper introduces a framework that combines a deep learning model with an attention mechanism that highlights the critical time steps in the forecasting process, thus improving model interpretability and supporting clinical decision-making. We show that the attention mechanism could be adapted to various black box time series forecasting models such as N-HiTS and N-BEATS. Our method preserves the accuracy of conventional deep learning models while enhancing interpretability through attention-weight-generated heatmaps. We evaluated our model on the eICU-CRD dataset, focusing on forecasting vital signs for sepsis patients. We assessed its performance using mean squared error (MSE) and dynamic time warping (DTW) metrics. We explored the attention maps of N-HiTS and N-BEATS, examining the differences in their performance and identifying crucial factors influencing vital sign forecasting.
SM70: A Large Language Model for Medical Devices
Dec 12, 2023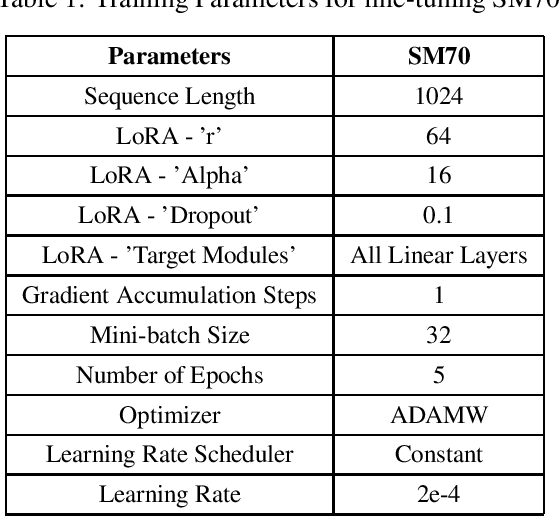
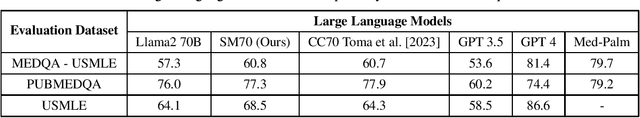
Abstract:We are introducing SM70, a 70 billion-parameter Large Language Model that is specifically designed for SpassMed's medical devices under the brand name 'JEE1' (pronounced as G1 and means 'Life'). This large language model provides more accurate and safe responses to medical-domain questions. To fine-tune SM70, we used around 800K data entries from the publicly available dataset MedAlpaca. The Llama2 70B open-sourced model served as the foundation for SM70, and we employed the QLoRA technique for fine-tuning. The evaluation is conducted across three benchmark datasets - MEDQA - USMLE, PUBMEDQA, and USMLE - each representing a unique aspect of medical knowledge and reasoning. The performance of SM70 is contrasted with other notable LLMs, including Llama2 70B, Clinical Camel 70 (CC70), GPT 3.5, GPT 4, and Med-Palm, to provide a comparative understanding of its capabilities within the medical domain. Our results indicate that SM70 outperforms several established models in these datasets, showcasing its proficiency in handling a range of medical queries, from fact-based questions derived from PubMed abstracts to complex clinical decision-making scenarios. The robust performance of SM70, particularly in the USMLE and PUBMEDQA datasets, suggests its potential as an effective tool in clinical decision support and medical information retrieval. Despite its promising results, the paper also acknowledges the areas where SM70 lags behind the most advanced model, GPT 4, thereby highlighting the need for further development, especially in tasks demanding extensive medical knowledge and intricate reasoning.
Vital Sign Forecasting for Sepsis Patients in ICUs
Nov 08, 2023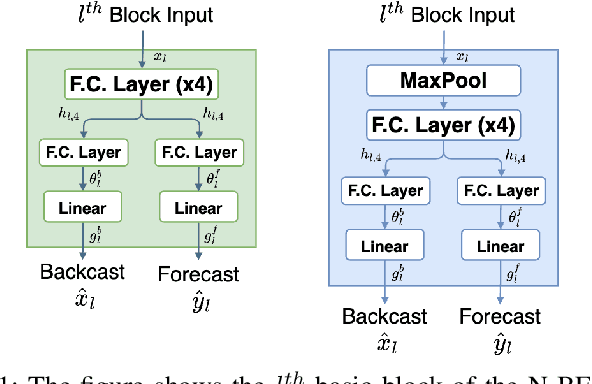
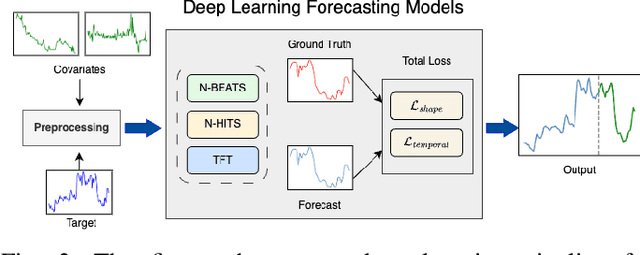
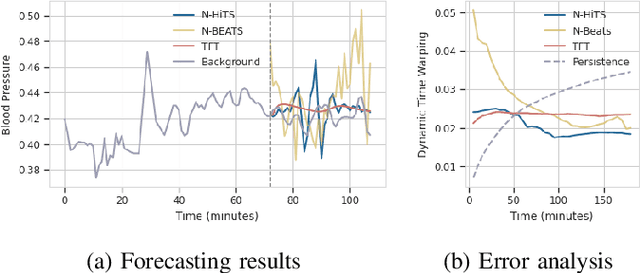
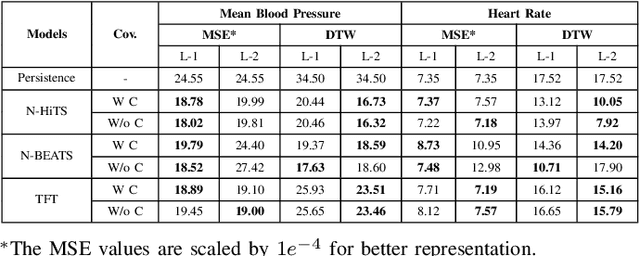
Abstract:Sepsis and septic shock are a critical medical condition affecting millions globally, with a substantial mortality rate. This paper uses state-of-the-art deep learning (DL) architectures to introduce a multi-step forecasting system to predict vital signs indicative of septic shock progression in Intensive Care Units (ICUs). Our approach utilizes a short window of historical vital sign data to forecast future physiological conditions. We introduce a DL-based vital sign forecasting system that predicts up to 3 hours of future vital signs from 6 hours of past data. We further adopt the DILATE loss function to capture better the shape and temporal dynamics of vital signs, which are critical for clinical decision-making. We compare three DL models, N-BEATS, N-HiTS, and Temporal Fusion Transformer (TFT), using the publicly available eICU Collaborative Research Database (eICU-CRD), highlighting their forecasting capabilities in a critical care setting. We evaluate the performance of our models using mean squared error (MSE) and dynamic time warping (DTW) metrics. Our findings show that while TFT excels in capturing overall trends, N-HiTS is superior in retaining short-term fluctuations within a predefined range. This paper demonstrates the potential of deep learning in transforming the monitoring systems in ICUs, potentially leading to significant improvements in patient care and outcomes by accurately forecasting vital signs to assist healthcare providers in detecting early signs of physiological instability and anticipating septic shock.
Extending Machine Learning-Based Early Sepsis Detection to Different Demographics
Nov 07, 2023Abstract:Sepsis requires urgent diagnosis, but research is predominantly focused on Western datasets. In this study, we perform a comparative analysis of two ensemble learning methods, LightGBM and XGBoost, using the public eICU-CRD dataset and a private South Korean St. Mary's Hospital's dataset. Our analysis reveals the effectiveness of these methods in addressing healthcare data imbalance and enhancing sepsis detection. Specifically, LightGBM shows a slight edge in computational efficiency and scalability. The study paves the way for the broader application of machine learning in critical care, thereby expanding the reach of predictive analytics in healthcare globally.
Interpreting Forecasted Vital Signs Using N-BEATS in Sepsis Patients
Jun 24, 2023

Abstract:Detecting and predicting septic shock early is crucial for the best possible outcome for patients. Accurately forecasting the vital signs of patients with sepsis provides valuable insights to clinicians for timely interventions, such as administering stabilizing drugs or optimizing infusion strategies. Our research examines N-BEATS, an interpretable deep-learning forecasting model that can forecast 3 hours of vital signs for sepsis patients in intensive care units (ICUs). In this work, we use the N-BEATS interpretable configuration to forecast the vital sign trends and compare them with the actual trend to understand better the patient's changing condition and the effects of infused drugs on their vital signs. We evaluate our approach using the publicly available eICU Collaborative Research Database dataset and rigorously evaluate the vital sign forecasts using out-of-sample evaluation criteria. We present the performance of our model using error metrics, including mean squared error (MSE), mean average percentage error (MAPE), and dynamic time warping (DTW), where the best scores achieved are 18.52e-4, 7.60, and 17.63e-3, respectively. We analyze the samples where the forecasted trend does not match the actual trend and study the impact of infused drugs on changing the actual vital signs compared to the forecasted trend. Additionally, we examined the mortality rates of patients where the actual trend and the forecasted trend did not match. We observed that the mortality rate was higher (92%) when the actual and forecasted trends closely matched, compared to when they were not similar (84%).
Investigating Poor Performance Regions of Black Boxes: LIME-based Exploration in Sepsis Detection
Jun 21, 2023
Abstract:Interpreting machine learning models remains a challenge, hindering their adoption in clinical settings. This paper proposes leveraging Local Interpretable Model-Agnostic Explanations (LIME) to provide interpretable descriptions of black box classification models in high-stakes sepsis detection. By analyzing misclassified instances, significant features contributing to suboptimal performance are identified. The analysis reveals regions where the classifier performs poorly, allowing the calculation of error rates within these regions. This knowledge is crucial for cautious decision-making in sepsis detection and other critical applications. The proposed approach is demonstrated using the eICU dataset, effectively identifying and visualizing regions where the classifier underperforms. By enhancing interpretability, our method promotes the adoption of machine learning models in clinical practice, empowering informed decision-making and mitigating risks in critical scenarios.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge