Surajsinh Parmar
Beyond LoRA: Exploring Efficient Fine-Tuning Techniques for Time Series Foundational Models
Sep 17, 2024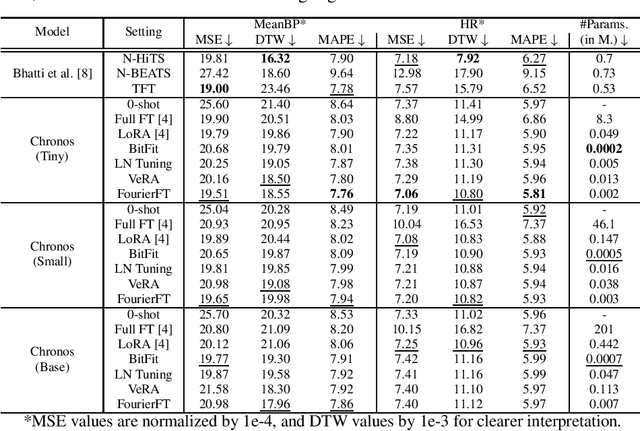
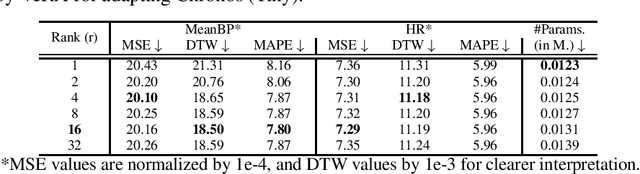
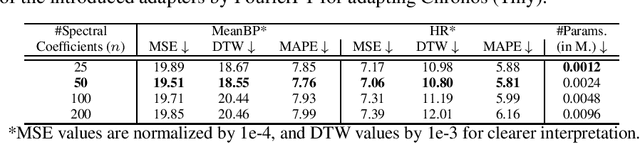

Abstract:Time Series Foundation Models (TSFMs) have recently garnered attention for their ability to model complex, large-scale time series data across domains such as retail, finance, and transportation. However, their application to sensitive, domain-specific fields like healthcare remains challenging, primarily due to the difficulty of fine-tuning these models for specialized, out-of-domain tasks with scarce publicly available datasets. In this work, we explore the use of Parameter-Efficient Fine-Tuning (PEFT) techniques to address these limitations, focusing on healthcare applications, particularly ICU vitals forecasting for sepsis patients. We introduce and evaluate two selective (BitFit and LayerNorm Tuning) and two additive (VeRA and FourierFT) PEFT techniques on multiple configurations of the Chronos TSFM for forecasting vital signs of sepsis patients. Our comparative analysis demonstrates that some of these PEFT methods outperform LoRA in terms of parameter efficiency and domain adaptation, establishing state-of-the-art (SOTA) results in ICU vital forecasting tasks. Interestingly, FourierFT applied to the Chronos (Tiny) variant surpasses the SOTA model while fine-tuning only 2,400 parameters compared to the 700K parameters of the benchmark.
Towards Democratizing Multilingual Large Language Models For Medicine Through A Two-Stage Instruction Fine-tuning Approach
Sep 09, 2024



Abstract:Open-source, multilingual medical large language models (LLMs) have the potential to serve linguistically diverse populations across different regions. Adapting generic LLMs for healthcare often requires continual pretraining, but this approach is computationally expensive and sometimes impractical. Instruction fine-tuning on a specific task may not always guarantee optimal performance due to the lack of broader domain knowledge that the model needs to understand and reason effectively in diverse scenarios. To address these challenges, we introduce two multilingual instruction fine-tuning datasets, MMed-IFT and MMed-IFT-MC, containing over 200k high-quality medical samples in six languages. We propose a two-stage training paradigm: the first stage injects general medical knowledge using MMed-IFT, while the second stage fine-tunes task-specific multiple-choice questions with MMed-IFT-MC. Our method achieves competitive results on both English and multilingual benchmarks, striking a balance between computational efficiency and performance. We plan to make our dataset and model weights public at \url{https://github.com/SpassMed/Med-Llama3} in the future.
SM70: A Large Language Model for Medical Devices
Dec 12, 2023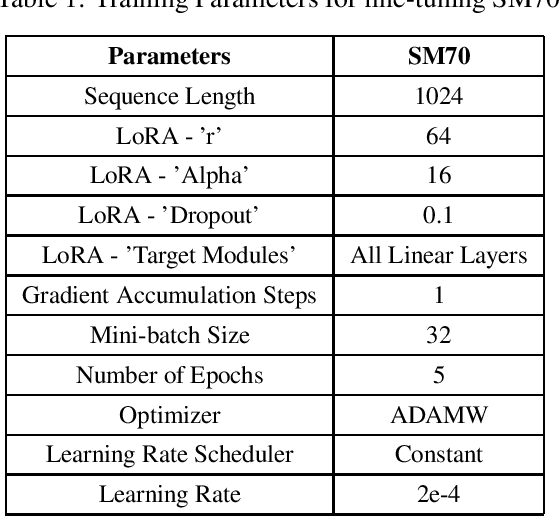
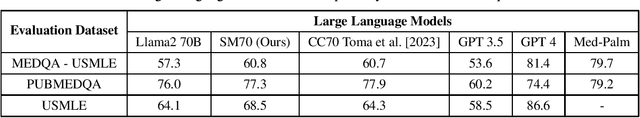
Abstract:We are introducing SM70, a 70 billion-parameter Large Language Model that is specifically designed for SpassMed's medical devices under the brand name 'JEE1' (pronounced as G1 and means 'Life'). This large language model provides more accurate and safe responses to medical-domain questions. To fine-tune SM70, we used around 800K data entries from the publicly available dataset MedAlpaca. The Llama2 70B open-sourced model served as the foundation for SM70, and we employed the QLoRA technique for fine-tuning. The evaluation is conducted across three benchmark datasets - MEDQA - USMLE, PUBMEDQA, and USMLE - each representing a unique aspect of medical knowledge and reasoning. The performance of SM70 is contrasted with other notable LLMs, including Llama2 70B, Clinical Camel 70 (CC70), GPT 3.5, GPT 4, and Med-Palm, to provide a comparative understanding of its capabilities within the medical domain. Our results indicate that SM70 outperforms several established models in these datasets, showcasing its proficiency in handling a range of medical queries, from fact-based questions derived from PubMed abstracts to complex clinical decision-making scenarios. The robust performance of SM70, particularly in the USMLE and PUBMEDQA datasets, suggests its potential as an effective tool in clinical decision support and medical information retrieval. Despite its promising results, the paper also acknowledges the areas where SM70 lags behind the most advanced model, GPT 4, thereby highlighting the need for further development, especially in tasks demanding extensive medical knowledge and intricate reasoning.
Extending Machine Learning-Based Early Sepsis Detection to Different Demographics
Nov 07, 2023Abstract:Sepsis requires urgent diagnosis, but research is predominantly focused on Western datasets. In this study, we perform a comparative analysis of two ensemble learning methods, LightGBM and XGBoost, using the public eICU-CRD dataset and a private South Korean St. Mary's Hospital's dataset. Our analysis reveals the effectiveness of these methods in addressing healthcare data imbalance and enhancing sepsis detection. Specifically, LightGBM shows a slight edge in computational efficiency and scalability. The study paves the way for the broader application of machine learning in critical care, thereby expanding the reach of predictive analytics in healthcare globally.
Investigating Poor Performance Regions of Black Boxes: LIME-based Exploration in Sepsis Detection
Jun 21, 2023
Abstract:Interpreting machine learning models remains a challenge, hindering their adoption in clinical settings. This paper proposes leveraging Local Interpretable Model-Agnostic Explanations (LIME) to provide interpretable descriptions of black box classification models in high-stakes sepsis detection. By analyzing misclassified instances, significant features contributing to suboptimal performance are identified. The analysis reveals regions where the classifier performs poorly, allowing the calculation of error rates within these regions. This knowledge is crucial for cautious decision-making in sepsis detection and other critical applications. The proposed approach is demonstrated using the eICU dataset, effectively identifying and visualizing regions where the classifier underperforms. By enhancing interpretability, our method promotes the adoption of machine learning models in clinical practice, empowering informed decision-making and mitigating risks in critical scenarios.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge