Cynthia Hawkins
Tumor Location-weighted MRI-Report Contrastive Learning: A Framework for Improving the Explainability of Pediatric Brain Tumor Diagnosis
Nov 01, 2024



Abstract:Despite the promising performance of convolutional neural networks (CNNs) in brain tumor diagnosis from magnetic resonance imaging (MRI), their integration into the clinical workflow has been limited. That is mainly due to the fact that the features contributing to a model's prediction are unclear to radiologists and hence, clinically irrelevant, i.e., lack of explainability. As the invaluable sources of radiologists' knowledge and expertise, radiology reports can be integrated with MRI in a contrastive learning (CL) framework, enabling learning from image-report associations, to improve CNN explainability. In this work, we train a multimodal CL architecture on 3D brain MRI scans and radiology reports to learn informative MRI representations. Furthermore, we integrate tumor location, salient to several brain tumor analysis tasks, into this framework to improve its generalizability. We then apply the learnt image representations to improve explainability and performance of genetic marker classification of pediatric Low-grade Glioma, the most prevalent brain tumor in children, as a downstream task. Our results indicate a Dice score of 31.1% between the model's attention maps and manual tumor segmentation (as an explainability measure) with test classification performance of 87.7%, significantly outperforming the baselines. These enhancements can build trust in our model among radiologists, facilitating its integration into clinical practices for more efficient tumor diagnosis.
Improving Pediatric Low-Grade Neuroepithelial Tumors Molecular Subtype Identification Using a Novel AUROC Loss Function for Convolutional Neural Networks
Feb 05, 2024

Abstract:Pediatric Low-Grade Neuroepithelial Tumors (PLGNT) are the most common pediatric cancer type, accounting for 40% of brain tumors in children, and identifying PLGNT molecular subtype is crucial for treatment planning. However, the gold standard to determine the PLGNT subtype is biopsy, which can be impractical or dangerous for patients. This research improves the performance of Convolutional Neural Networks (CNNs) in classifying PLGNT subtypes through MRI scans by introducing a loss function that specifically improves the model's Area Under the Receiver Operating Characteristic (ROC) Curve (AUROC), offering a non-invasive diagnostic alternative. In this study, a retrospective dataset of 339 children with PLGNT (143 BRAF fusion, 71 with BRAF V600E mutation, and 125 non-BRAF) was curated. We employed a CNN model with Monte Carlo random data splitting. The baseline model was trained using binary cross entropy (BCE), and achieved an AUROC of 86.11% for differentiating BRAF fusion and BRAF V600E mutations, which was improved to 87.71% using our proposed AUROC loss function (p-value 0.045). With multiclass classification, the AUROC improved from 74.42% to 76. 59% (p-value 0.0016).
Generating 3D Brain Tumor Regions in MRI using Vector-Quantization Generative Adversarial Networks
Oct 02, 2023



Abstract:Medical image analysis has significantly benefited from advancements in deep learning, particularly in the application of Generative Adversarial Networks (GANs) for generating realistic and diverse images that can augment training datasets. However, the effectiveness of such approaches is often limited by the amount of available data in clinical settings. Additionally, the common GAN-based approach is to generate entire image volumes, rather than solely the region of interest (ROI). Research on deep learning-based brain tumor classification using MRI has shown that it is easier to classify the tumor ROIs compared to the entire image volumes. In this work, we present a novel framework that uses vector-quantization GAN and a transformer incorporating masked token modeling to generate high-resolution and diverse 3D brain tumor ROIs that can be directly used as augmented data for the classification of brain tumor ROI. We apply our method to two imbalanced datasets where we augment the minority class: (1) the Multimodal Brain Tumor Segmentation Challenge (BraTS) 2019 dataset to generate new low-grade glioma (LGG) ROIs to balance with high-grade glioma (HGG) class; (2) the internal pediatric LGG (pLGG) dataset tumor ROIs with BRAF V600E Mutation genetic marker to balance with BRAF Fusion genetic marker class. We show that the proposed method outperforms various baseline models in both qualitative and quantitative measurements. The generated data was used to balance the data in the brain tumor types classification task. Using the augmented data, our approach surpasses baseline models by 6.4% in AUC on the BraTS 2019 dataset and 4.3% in AUC on our internal pLGG dataset. The results indicate the generated tumor ROIs can effectively address the imbalanced data problem. Our proposed method has the potential to facilitate an accurate diagnosis of rare brain tumors using MRI scans.
A novel GAN-based paradigm for weakly supervised brain tumor segmentation of MR images
Nov 10, 2022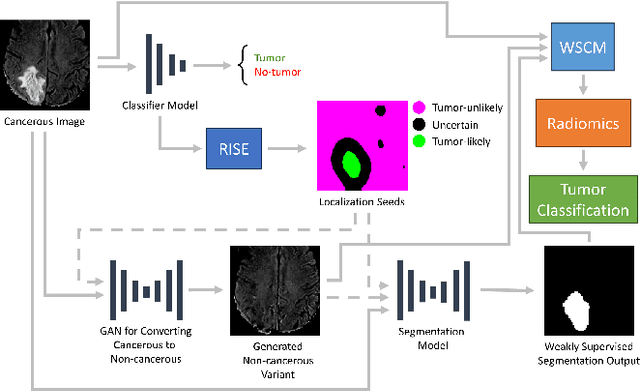
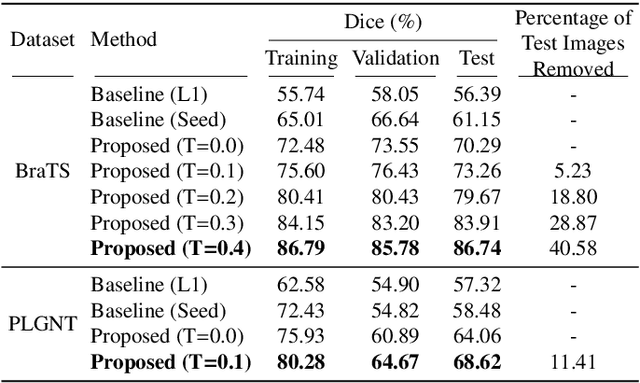
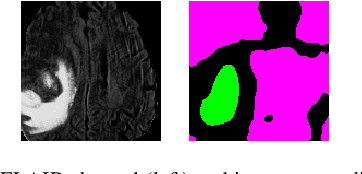
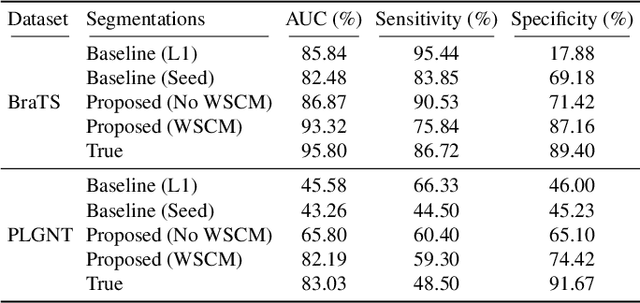
Abstract:Segmentation of regions of interest (ROIs) for identifying abnormalities is a leading problem in medical imaging. Using Machine Learning (ML) for this problem generally requires manually annotated ground-truth segmentations, demanding extensive time and resources from radiologists. This work presents a novel weakly supervised approach that utilizes binary image-level labels, which are much simpler to acquire, to effectively segment anomalies in medical Magnetic Resonance (MR) images without ground truth annotations. We train a binary classifier using these labels and use it to derive seeds indicating regions likely and unlikely to contain tumors. These seeds are used to train a generative adversarial network (GAN) that converts cancerous images to healthy variants, which are then used in conjunction with the seeds to train a ML model that generates effective segmentations. This method produces segmentations that achieve Dice coefficients of 0.7903, 0.7868, and 0.7712 on the MICCAI Brain Tumor Segmentation (BraTS) 2020 dataset for the training, validation, and test cohorts respectively. We also propose a weakly supervised means of filtering the segmentations, removing a small subset of poorer segmentations to acquire a large subset of high quality segmentations. The proposed filtering further improves the Dice coefficients to up to 0.8374, 0.8232, and 0.8136 for training, validation, and test, respectively.
Tumor-location-guided CNNs for Pediatric Low-grade Glioma Molecular Biomarker Classification Using MRI
Oct 13, 2022


Abstract:Pediatric low-grade glioma (pLGG) is the most common type of brain cancer among children, and the identification of molecular markers for pLGG is crucial for successful treatment planning. Current standard care is biopsy, which is invasive. Thus, the non-invasive imaging-based approaches, where Machine Learning (ML) has a high potential, are impactful. Recently, we developed a tumor-location-based algorithm and demonstrated its potential to differentiate pLGG molecular subtypes. In this work, we first reevaluated the performance of the location-based algorithm on a larger pLGG dataset, which includes 214 patients and achieved an area under the receiver operating characteristic curve (AUROC) of 77.90. A Convolutional Neural Network (CNN) based algorithm increased the average AUROC to 86.11. Ultimately, we designed and implemented a tumor-location-guided CNN algorithm and achieved average AUROC of 88.64. Using a repeated experiment approach with 100 runs, we ensured the results were reproducible and the improvement was statistically significant.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge