A novel GAN-based paradigm for weakly supervised brain tumor segmentation of MR images
Paper and Code
Nov 10, 2022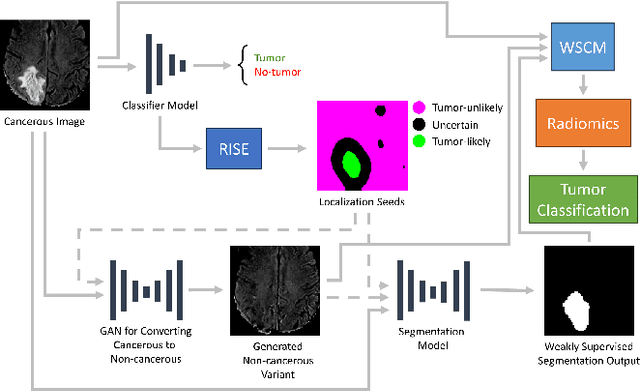
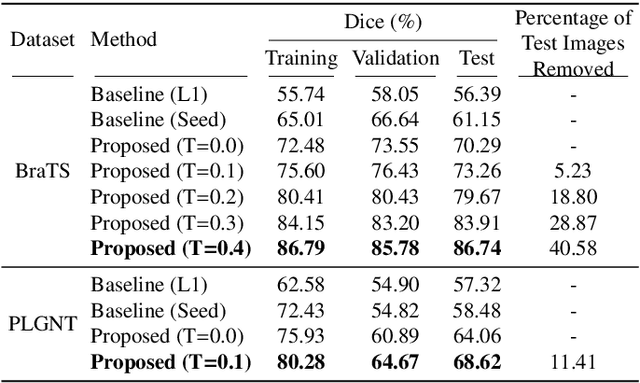
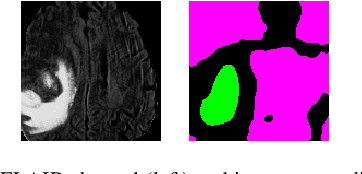
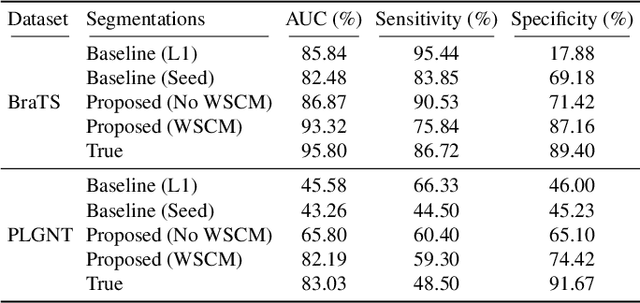
Segmentation of regions of interest (ROIs) for identifying abnormalities is a leading problem in medical imaging. Using Machine Learning (ML) for this problem generally requires manually annotated ground-truth segmentations, demanding extensive time and resources from radiologists. This work presents a novel weakly supervised approach that utilizes binary image-level labels, which are much simpler to acquire, to effectively segment anomalies in medical Magnetic Resonance (MR) images without ground truth annotations. We train a binary classifier using these labels and use it to derive seeds indicating regions likely and unlikely to contain tumors. These seeds are used to train a generative adversarial network (GAN) that converts cancerous images to healthy variants, which are then used in conjunction with the seeds to train a ML model that generates effective segmentations. This method produces segmentations that achieve Dice coefficients of 0.7903, 0.7868, and 0.7712 on the MICCAI Brain Tumor Segmentation (BraTS) 2020 dataset for the training, validation, and test cohorts respectively. We also propose a weakly supervised means of filtering the segmentations, removing a small subset of poorer segmentations to acquire a large subset of high quality segmentations. The proposed filtering further improves the Dice coefficients to up to 0.8374, 0.8232, and 0.8136 for training, validation, and test, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge