Jay J. Yoo
A Comprehensive Study of Radiomics-based Machine Learning for Fibrosis Detection
Nov 25, 2022Abstract:Objectives: Early detection of liver fibrosis can help cure the disease or prevent disease progression. We perform a comprehensive study of machine learning-based fibrosis detection in CT images using radiomic features to develop a non-invasive approach to fibrosis detection. Methods: Two sets of radiomic features were extracted from spherical ROIs in CT images of 182 patients who underwent simultaneous liver biopsy and CT examinations, one set corresponding to biopsy locations and another distant from biopsy locations. Combinations of contrast, normalization, machine learning model, feature selection method, bin width, and kernel radius were investigated, each of which were trained and evaluated 100 times with randomized development and test cohorts. The best settings were evaluated based on their mean test AUC and the best features were determined based on their frequency among the best settings. Results: Logistic regression models with NC images normalized using Gamma correction with $\gamma = 1.5$ performed best for fibrosis detection. Boruta was the best for radiomic feature selection method. Training a model using these optimal settings and features consisting of first order energy, first order kurtosis, and first order skewness, resulted in a model that achieved mean test AUCs of 0.7549 and 0.7166 on biopsy-based and non-biopsy ROIs respectively, outperforming a baseline and best models found during the initial study. Conclusions: Logistic regression models trained on radiomic features from NC images normalized using Gamma correction with $\gamma = 1.5$ that underwent Boruta feature selection are effective for liver fibrosis detection. Energy, kurtosis, and skewness are particularly effective features for fibrosis detection.
A novel GAN-based paradigm for weakly supervised brain tumor segmentation of MR images
Nov 10, 2022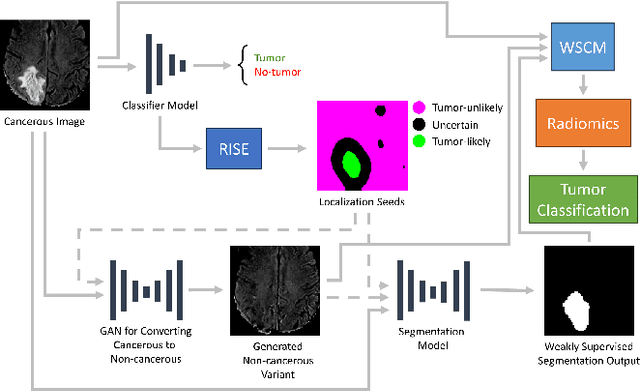
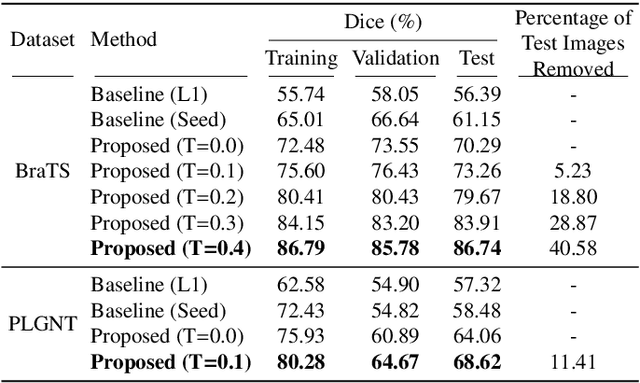
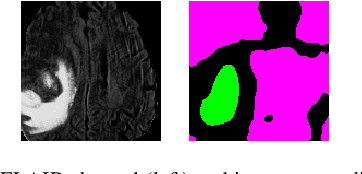
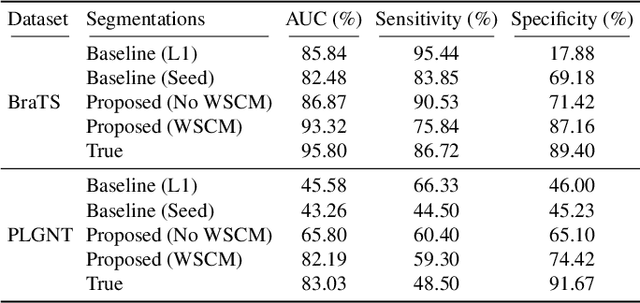
Abstract:Segmentation of regions of interest (ROIs) for identifying abnormalities is a leading problem in medical imaging. Using Machine Learning (ML) for this problem generally requires manually annotated ground-truth segmentations, demanding extensive time and resources from radiologists. This work presents a novel weakly supervised approach that utilizes binary image-level labels, which are much simpler to acquire, to effectively segment anomalies in medical Magnetic Resonance (MR) images without ground truth annotations. We train a binary classifier using these labels and use it to derive seeds indicating regions likely and unlikely to contain tumors. These seeds are used to train a generative adversarial network (GAN) that converts cancerous images to healthy variants, which are then used in conjunction with the seeds to train a ML model that generates effective segmentations. This method produces segmentations that achieve Dice coefficients of 0.7903, 0.7868, and 0.7712 on the MICCAI Brain Tumor Segmentation (BraTS) 2020 dataset for the training, validation, and test cohorts respectively. We also propose a weakly supervised means of filtering the segmentations, removing a small subset of poorer segmentations to acquire a large subset of high quality segmentations. The proposed filtering further improves the Dice coefficients to up to 0.8374, 0.8232, and 0.8136 for training, validation, and test, respectively.
Superpixel Generation and Clustering for Weakly Supervised Brain Tumor Segmentation in MR Images
Sep 20, 2022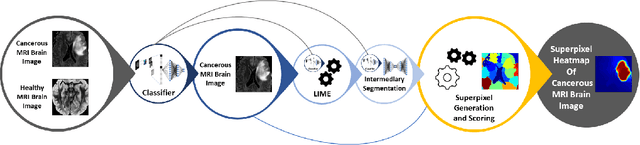
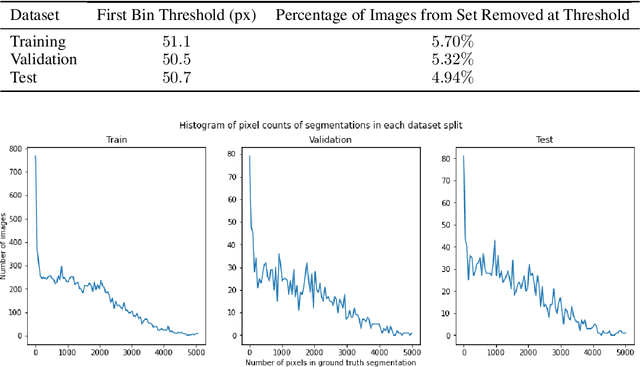
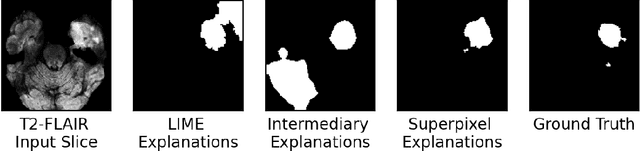
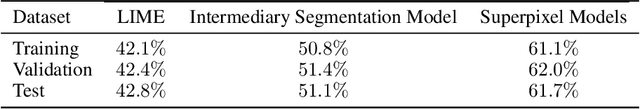
Abstract:Training Machine Learning (ML) models to segment tumors and other anomalies in medical images is an increasingly popular area of research but generally requires manually annotated ground truth segmentations which necessitates significant time and resources to create. This work proposes a pipeline of ML models that utilize binary classification labels, which can be easily acquired, to segment ROIs without requiring ground truth annotations. We used 2D slices of Magnetic Resonance Imaging (MRI) brain scans from the Multimodal Brain Tumor Segmentation Challenge (BraTS) 2020 dataset and labels indicating the presence of high-grade glioma (HGG) tumors to train the pipeline. Our pipeline also introduces a novel variation of deep learning-based superpixel generation, which enables training guided by clustered superpixels and simultaneously trains a superpixel clustering model. On our test set, our pipeline's segmentations achieved a Dice coefficient of 61.7%, which is a substantial improvement over the 42.8% Dice coefficient acquired when the popular Local Interpretable Model-Agnostic Explanations (LIME) method was used.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge