Superpixel Generation and Clustering for Weakly Supervised Brain Tumor Segmentation in MR Images
Paper and Code
Sep 20, 2022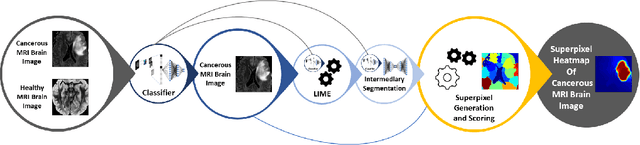
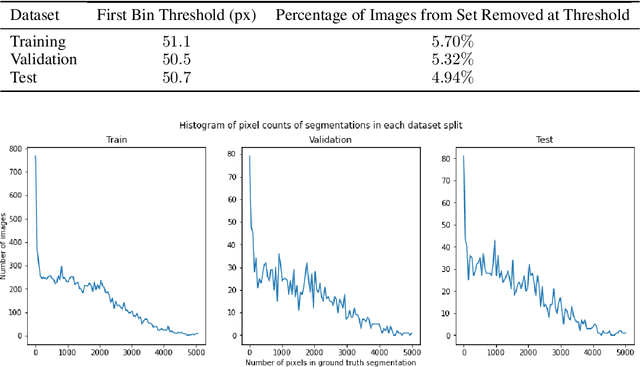
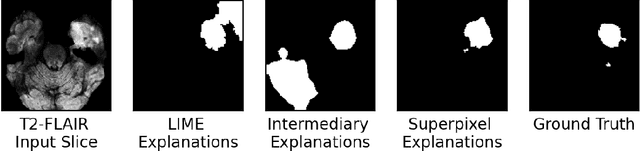
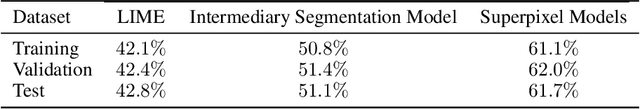
Training Machine Learning (ML) models to segment tumors and other anomalies in medical images is an increasingly popular area of research but generally requires manually annotated ground truth segmentations which necessitates significant time and resources to create. This work proposes a pipeline of ML models that utilize binary classification labels, which can be easily acquired, to segment ROIs without requiring ground truth annotations. We used 2D slices of Magnetic Resonance Imaging (MRI) brain scans from the Multimodal Brain Tumor Segmentation Challenge (BraTS) 2020 dataset and labels indicating the presence of high-grade glioma (HGG) tumors to train the pipeline. Our pipeline also introduces a novel variation of deep learning-based superpixel generation, which enables training guided by clustered superpixels and simultaneously trains a superpixel clustering model. On our test set, our pipeline's segmentations achieved a Dice coefficient of 61.7%, which is a substantial improvement over the 42.8% Dice coefficient acquired when the popular Local Interpretable Model-Agnostic Explanations (LIME) method was used.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge